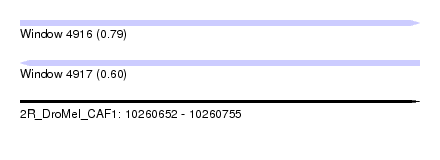
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,260,652 – 10,260,755 |
| Length | 103 |
| Max. P | 0.789718 |

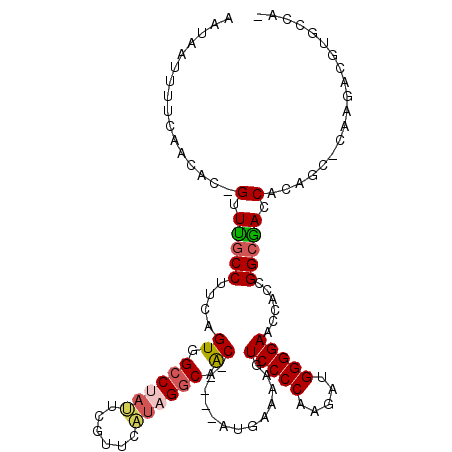
| Location | 10,260,652 – 10,260,755 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -32.92 |
| Consensus MFE | -21.86 |
| Energy contribution | -22.03 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.789718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10260652 103 + 20766785 AAUAAUUUUCAACAC-GUUUGCCUUCAGUCGCCUAUUCGUUGAUAGGCAAC-----AUGAAAAGUCCCCAAGAUGGGGAAUCACCGGCAACCACAGC-CAAGACGUGCCA- ............(((-((((...((((((.(((((((....))))))).))-----.))))...(((((.....)))))......(((.......))-).)))))))...- ( -32.60) >DroSec_CAF1 4470 103 + 1 AAUAAUUUCCAACAC-GUUUGCCUUCAGUGGCCUACUCGUUCGUAGGCAAC-----AAGAAAAGUCCCCAAGAUGGGGAACCACCGGCGACCACAGC-CAAGACGUGCCA- ............(((-((((.......(((((((((......))))))...-----........(((((.....)))))..))).(((.......))-).)))))))...- ( -33.90) >DroSim_CAF1 4372 103 + 1 AAUAAUUUCCAACAC-GUUUGCCUUCAGUGGCCUACUCGUUCGUAGGCAAC-----AAGAAAAGUCCCCAAGAUGGGGAACCACCGGCGACCGCAGC-CAAGACGUGCCA- ............(((-((((.......(((((((((......))))))...-----........(((((.....)))))..))).(((.......))-).)))))))...- ( -33.90) >DroEre_CAF1 6353 103 + 1 AAUAAUUUUCAACAC-GUUUGCCUUCAGUGGCCUAUGCGGUGAUAGGCGAC-----AUGAAAAGUCCCCAAGAUGGGGAACCACCGGCGACCACAGC-CACGACGUGCCU- ............(((-(((....((((((.((((((......)))))).))-----.))))...(((((.....)))))......(((.......))-)..))))))...- ( -33.80) >DroYak_CAF1 4546 103 + 1 AAUAAAGUUCAACUC-GUUUGCCUUCAGUGGCCAAUGCGCUUAUAGGCGGC-----AUGAAAAGUCCCCAAGAUGGGGAACCACCGGCAACCGCAGC-CACGACGUGCCU- ...............-....((((..((((.......))))...))))(((-----(((.....(((((.....)))))......(((.......))-)....)))))).- ( -32.40) >DroAna_CAF1 4918 107 + 1 GCAAAUUGUCGUCGCAGUUCACCUUCGGUGGCCAAUUUAUU----GUCAACACCAGAUGAAAAGUCCCCGCUCUGGGGAACCACCGGCGACCAGACCACAAGACGUGACUG .......(((((((..((((.(((..(((((...((((.(.----.((.......))..).))))..)))))..)))))))...)))))))(((..(((.....))).))) ( -30.90) >consensus AAUAAUUUUCAACAC_GUUUGCCUUCAGUGGCCUAUUCGUUCAUAGGCAAC_____AUGAAAAGUCCCCAAGAUGGGGAACCACCGGCGACCACAGC_CAAGACGUGCCA_ ................(.(((((....((.((((((......)))))).)).............(((((.....)))))......))))).)................... (-21.86 = -22.03 + 0.17)


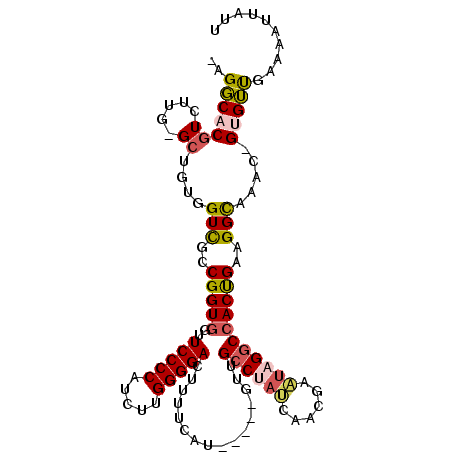
| Location | 10,260,652 – 10,260,755 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.51 |
| Mean single sequence MFE | -37.16 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.603544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10260652 103 - 20766785 -UGGCACGUCUUG-GCUGUGGUUGCCGGUGAUUCCCCAUCUUGGGGACUUUUCAU-----GUUGCCUAUCAACGAAUAGGCGACUGAAGGCAAAC-GUGUUGAAAAUUAUU -.(((((((.(((-.((.((((((((.((((.(((((.....)))))....))))-----((((.....)))).....)))))))).)).)))))-))))).......... ( -38.40) >DroSec_CAF1 4470 103 - 1 -UGGCACGUCUUG-GCUGUGGUCGCCGGUGGUUCCCCAUCUUGGGGACUUUUCUU-----GUUGCCUACGAACGAGUAGGCCACUGAAGGCAAAC-GUGUUGGAAAUUAUU -.(((((((...(-((....)))(((...(((((((......))))))).(((..-----((.((((((......)))))).)).))))))..))-))))).......... ( -37.20) >DroSim_CAF1 4372 103 - 1 -UGGCACGUCUUG-GCUGCGGUCGCCGGUGGUUCCCCAUCUUGGGGACUUUUCUU-----GUUGCCUACGAACGAGUAGGCCACUGAAGGCAAAC-GUGUUGGAAAUUAUU -.(((((((.(((-.((((....))(((((..(((((.....)))))........-----...((((((......))))))))))).)).)))))-))))).......... ( -37.90) >DroEre_CAF1 6353 103 - 1 -AGGCACGUCGUG-GCUGUGGUCGCCGGUGGUUCCCCAUCUUGGGGACUUUUCAU-----GUCGCCUAUCACCGCAUAGGCCACUGAAGGCAAAC-GUGUUGAAAAUUAUU -.(((((((..((-.((((((((..((((((((((((.....)))))........-----.......)))))))....))))))...)).)).))-))))).......... ( -36.66) >DroYak_CAF1 4546 103 - 1 -AGGCACGUCGUG-GCUGCGGUUGCCGGUGGUUCCCCAUCUUGGGGACUUUUCAU-----GCCGCCUAUAAGCGCAUUGGCCACUGAAGGCAAAC-GAGUUGAACUUUAUU -..((...(((((-((((((.(((..(((((((((((.....)))))........-----))))))..))).)))...)))))).))..))....-............... ( -37.40) >DroAna_CAF1 4918 107 - 1 CAGUCACGUCUUGUGGUCUGGUCGCCGGUGGUUCCCCAGAGCGGGGACUUUUCAUCUGGUGUUGAC----AAUAAAUUGGCCACCGAAGGUGAACUGCGACGACAAUUUGC (((((((.....)))).)))(((((((((((((((((.....)))))....(((........))).----.........)))))))..((....))))))).......... ( -35.40) >consensus _AGGCACGUCUUG_GCUGUGGUCGCCGGUGGUUCCCCAUCUUGGGGACUUUUCAU_____GUUGCCUAUCAACGAAUAGGCCACUGAAGGCAAAC_GUGUUGAAAAUUAUU ..(((((((.....))....(((..(((((..(((((.....)))))................((((((......)))))))))))..))).....))))).......... (-23.86 = -24.12 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:22 2006