
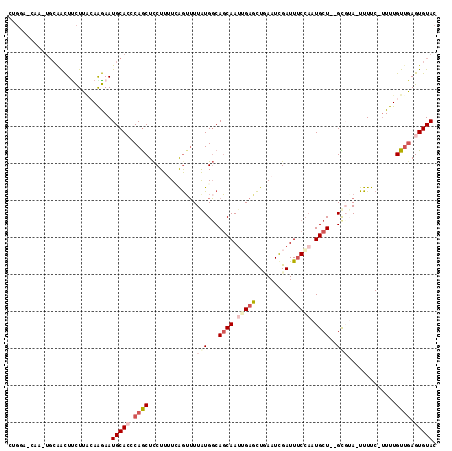
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,241,010 – 10,241,117 |
| Length | 107 |
| Max. P | 0.997294 |

| Location | 10,241,010 – 10,241,117 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 75.66 |
| Mean single sequence MFE | -28.76 |
| Consensus MFE | -7.61 |
| Energy contribution | -9.69 |
| Covariance contribution | 2.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.26 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
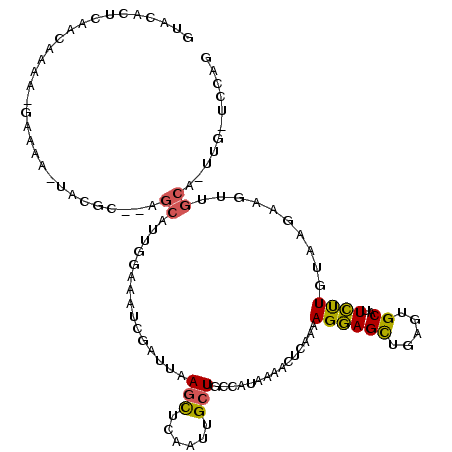
>2R_DroMel_CAF1 10241010 107 + 20766785 CUGCA--------AACUCCUUGCAAGAAUGCACACAGCUCCUUUUCAAAUUUAUGGCAUCAAUUGAGUUUAAUCGAUUUCCAAUGCUGUGCGUA-UUUUCCCUUUGUUAAGUGUAC .((((--------(.....)))))(((((((.((((((.((.............))....((((((......))))))......)))))).)))-))))................. ( -21.72) >DroSec_CAF1 69816 112 + 1 CUGGAGCAAGUGCAACUUCUUACAAGAAUGCAACCAGCUCCUUUUUAGUUUUAUGGCAGCAAAGGAGCUCAAUCGAUUUCCAAUGCU--GCCUA-UUUUC-UUUUGUUGAGUGUAC ..(((((...((((..((((....))))))))....))))).............(((((((..((((.((....)).))))..))))--)))((-(.(((-.......))).))). ( -32.70) >DroSim_CAF1 71790 112 + 1 CUGGAGCAAUUGCAACUUCUUACAAGAAUGCAACCAGCUCCUUUUCAGUUUUGUGGCAGCAAUGGAGCUUAAUCGAUUUCCAAUGCU--GCCUA-UUUUC-UUUUGUUGAGUGUAC ..(((((..(((((..((((....)))))))))...))))).............(((((((.(((((.((....)).))))).))))--)))((-(.(((-.......))).))). ( -33.00) >DroEre_CAF1 74604 112 + 1 CUGGCCCAAGUGCAACUCCUUAGAGGAAUGCACUCAACUCCUUAUGGGUUUUAUGGCAGCAAUUGAACUGAAUUGAUUGCC-AUGCU--GUGGACUUUUC-UUUUGUCGAGUGUAC ..((((((((((((..(((.....))).))))))..........)))))).(((((((((((((......))))).)))))-)))..--(((.((((..(-....)..)))).))) ( -32.90) >DroYak_CAF1 71640 103 + 1 ----------UGCGGUUUUUUUGAGAAAUGCACUCAGCUUCUUAUGGGUAUUAUGUCAGCAAUUGAACUGAAUCGAUUUCGAAUGCU--GUGGAAUUUUC-UUUUGUUGAGUGUAC ----------..................((((((((((.......(((.(((....(((((.(((((..........))))).))))--)...))).)))-....)))))))))). ( -23.50) >consensus CUGGA_CAA_UGCAACUUCUUACAAGAAUGCACCCAGCUCCUUUUCAGUUUUAUGGCAGCAAUUGAGCUGAAUCGAUUUCCAAUGCU__GCGUA_UUUUC_UUUUGUUGAGUGUAC ............................(((((.((((.............(((...((((.(((((..........))))).))))..))).............)))).))))). ( -7.61 = -9.69 + 2.08)


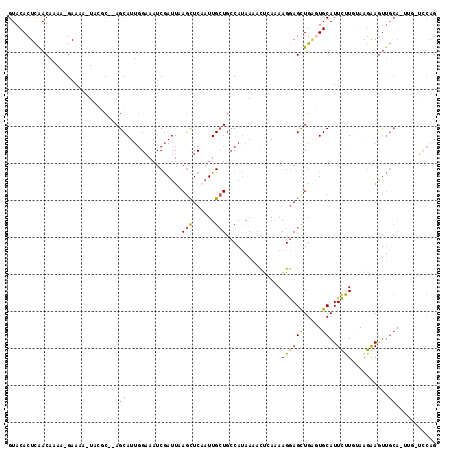
| Location | 10,241,010 – 10,241,117 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 75.66 |
| Mean single sequence MFE | -29.60 |
| Consensus MFE | -3.94 |
| Energy contribution | -3.58 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.13 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10241010 107 - 20766785 GUACACUUAACAAAGGGAAAA-UACGCACAGCAUUGGAAAUCGAUUAAACUCAAUUGAUGCCAUAAAUUUGAAAAGGAGCUGUGUGCAUUCUUGCAAGGAGUU--------UGCAG (((.((((......(((((..-..((((((((..(((..(((((((......))))))).)))....(((....))).))))))))..))))).....)))).--------))).. ( -27.40) >DroSec_CAF1 69816 112 - 1 GUACACUCAACAAAA-GAAAA-UAGGC--AGCAUUGGAAAUCGAUUGAGCUCCUUUGCUGCCAUAAAACUAAAAAGGAGCUGGUUGCAUUCUUGUAAGAAGUUGCACUUGCUCCAG ...............-.....-..(((--((((..(((..((....))..)))..))))))).............(((((.(((.(((((((....))))..)))))).))))).. ( -36.80) >DroSim_CAF1 71790 112 - 1 GUACACUCAACAAAA-GAAAA-UAGGC--AGCAUUGGAAAUCGAUUAAGCUCCAUUGCUGCCACAAAACUGAAAAGGAGCUGGUUGCAUUCUUGUAAGAAGUUGCAAUUGCUCCAG ...............-.....-..(((--((((.((((............)))).))))))).............(((((.(((((((((((....))))..)))))))))))).. ( -36.10) >DroEre_CAF1 74604 112 - 1 GUACACUCGACAAAA-GAAAAGUCCAC--AGCAU-GGCAAUCAAUUCAGUUCAAUUGCUGCCAUAAAACCCAUAAGGAGUUGAGUGCAUUCCUCUAAGGAGUUGCACUUGGGCCAG ...............-.....((((((--(((((-((((..(((((......))))).)))))).....((....)).))))((((((((((.....)))).)))))))))))... ( -31.80) >DroYak_CAF1 71640 103 - 1 GUACACUCAACAAAA-GAAAAUUCCAC--AGCAUUCGAAAUCGAUUCAGUUCAAUUGCUGACAUAAUACCCAUAAGAAGCUGAGUGCAUUUCUCAAAAAAACCGCA---------- ((((..((.......-))........(--(((..(((....))).(((((......))))).................)))).))))...................---------- ( -15.90) >consensus GUACACUCAACAAAA_GAAAA_UACGC__AGCAUUGGAAAUCGAUUAAGCUCAAUUGCUGCCAUAAAACUCAAAAGGAGCUGAGUGCAUUCUUGUAAGAAGUUGCA_UUG_UCCAG ..............................((...............(((......)))...............((((((.....))..))))..........))........... ( -3.94 = -3.58 + -0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:16 2006