
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
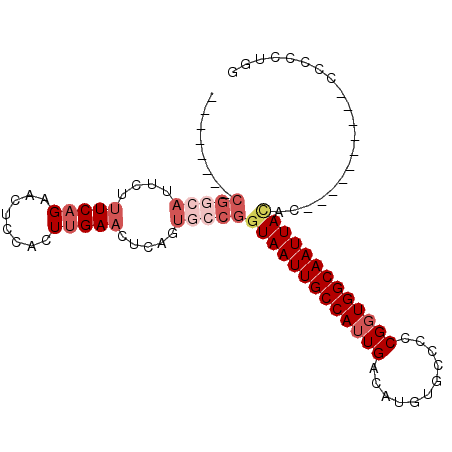
| Location | 10,229,665 – 10,229,793 |
| Length | 128 |
| Max. P | 0.979862 |

| Location | 10,229,665 – 10,229,759 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 77.21 |
| Mean single sequence MFE | -25.09 |
| Consensus MFE | -18.35 |
| Energy contribution | -20.02 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.979862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
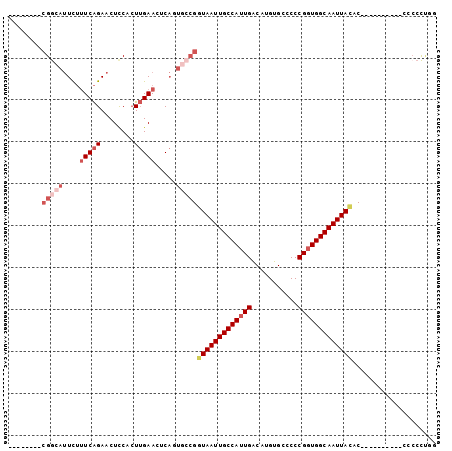
>2R_DroMel_CAF1 10229665 94 + 20766785 --------CGGCAUUCUUUCAGAACUCCACUUGAACUCACUGCCGGUAAUUGCCAUUGACAUGUGCUCCCGGUGGCAAUUAUACCACCCCCACCCCCCCUGG --------(((((....(((((........))))).....)))))(((((((((((((...........))))))))))))).................... ( -24.40) >DroSec_CAF1 58714 84 + 1 --------CGGCAUUCUUUCAGAACUCCACUUGAACUCAGUGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC----------CCCCCUGG --------(((((((..(((((........)))))...)))))))(((((((((((((...........)))))))))))))..----------........ ( -28.90) >DroSim_CAF1 58995 84 + 1 --------CGGCAUUCUUUCAGAACUCCACUUGAACUCAGUGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC----------CCCCCUGG --------(((((((..(((((........)))))...)))))))(((((((((((((...........)))))))))))))..----------........ ( -28.90) >DroEre_CAF1 63435 92 + 1 GUCCGGUCCGGCAUUCUUUCAGAACUCCACUUGAACUCACUUCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUAUAC----------CCCCCCGC ...(((.((((......(((((........))))).......))))((((((((((((...........))))))))))))...----------....))). ( -22.92) >DroYak_CAF1 58835 92 + 1 CUUCGCUUCGACAUUCUUUCAGAACUCCACUUGAACUCAGUGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC----------CCCCUCCC ........((.((((..(((((........)))))...)))).))(((((((((((((...........)))))))))))))..----------........ ( -21.90) >DroPer_CAF1 41500 72 + 1 --------GGGGAGUUGAUCGGAACUUCAGUGGAAAAC-------UUAAUUGCCACUGGCAUGUGCCCCCGAUGGCAAUUAC---------------CCGGG --------(((.((((((((((......(((((.((..-------....)).)))))(((....))).)))))..))))).)---------------))... ( -23.50) >consensus ________CGGCAUUCUUUCAGAACUCCACUUGAACUCAGUGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC__________CCCCCUGG ........(((((....(((((........))))).....)))))(((((((((((((...........))))))))))))).................... (-18.35 = -20.02 + 1.67)

| Location | 10,229,697 – 10,229,793 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 70.66 |
| Mean single sequence MFE | -25.95 |
| Consensus MFE | -17.58 |
| Energy contribution | -17.75 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.891119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
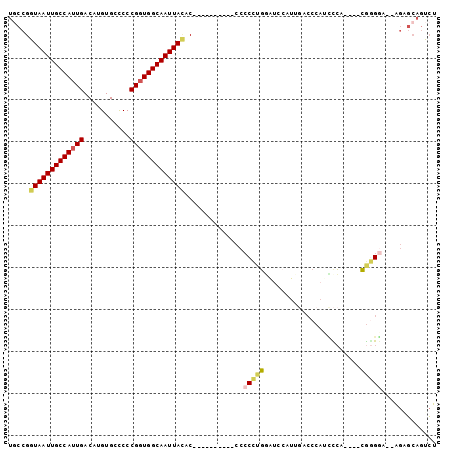
>2R_DroMel_CAF1 10229697 96 + 20766785 UGCCGGUAAUUGCCAUUGACAUGUGCUCCCGGUGGCAAUUAUACCACCCCCACCCCCCCUGGAUCCAUUGUCCCAUCCCU----CGGGGA--AGCGCAGUCU ....(((....)))...(((.((((((...(((((........))))).....((((...((((..........))))..----.)))).--))))))))). ( -30.80) >DroPse_CAF1 42443 77 + 1 -----CUAAUUGCCACUGACAUGUGCCCCCGAUGGCAAUUAC---------------CCGGGGCCCAUACACC-ACCUCA----CAGGACUCGAUCCACGGU -----......(((......(((.(((((....((......)---------------).))))).))).....-......----..(((.....)))..))) ( -17.20) >DroSec_CAF1 58746 90 + 1 UGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC----------CCCCCUGGAUCCAUUGACCCAUUCCAUUGACGGGGA--AGUGCAGUCU ((((.(((((((((((((...........))))))))))))).(----------(((..((((.............)))).....)))).--.).))).... ( -29.02) >DroSim_CAF1 59027 86 + 1 UGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC----------CCCCCUGGAUCCAUUGACCCAUCCCA----CGGGGA--AGAGCAGUCU ((((.(((((((((((((...........)))))))))))))..----------((((.(((...............)))----.)))).--.).))).... ( -29.96) >DroEre_CAF1 63475 86 + 1 UUCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUAUAC----------CCCCCCGCAUUGAUGGACCCAACCCA----CGAGGA--AGAGCAGUCU ((((.(((((((((((((...........)))))))))))))..----------.........(((.(((.......)))----))))))--)......... ( -22.20) >DroYak_CAF1 58875 75 + 1 UGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC----------CCCCUCCC-----------CCGGUGA----GGAGCA--GGAGCAGUCU (((..(((((((((((((...........))))))))))))).(----------(..((((.-----------.......----))))..--)).))).... ( -26.50) >consensus UGCCGGUAAUUGCCAUUGACAUGUGCCCCCGGUGGCAAUUACAC__________CCCCCUGGAUCCAUUGACCCAUCCCA____CGGGGA__AGAGCAGUCU .....(((((((((((((...........)))))))))))))..............(((((.......................)))))............. (-17.58 = -17.75 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:13 2006