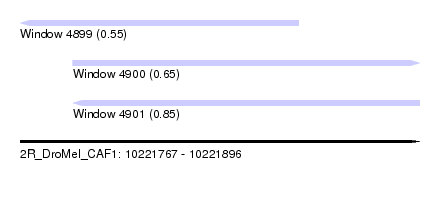
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,221,767 – 10,221,896 |
| Length | 129 |
| Max. P | 0.851375 |

| Location | 10,221,767 – 10,221,857 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 87.24 |
| Mean single sequence MFE | -19.20 |
| Consensus MFE | -15.01 |
| Energy contribution | -14.73 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
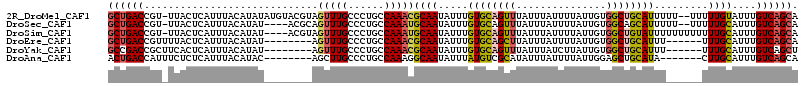
>2R_DroMel_CAF1 10221767 90 - 20766785 CCCUGCCAAACGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCUGCAUUUUU--UUUUUGUAUUUGUCAGCACAAAUGACUAGCUUUUG ..(((.((((..(((.....((((((((...............))))))))....--...)))..)))).)))................... ( -16.96) >DroSec_CAF1 50965 90 - 1 CCCUGCCAAAUGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCAGCAUUUUU--UUUUUGCAUUUGUCAGCAGAAAUGACUGCCUUUUG ..((((((((((...)))))).)))).................(((((((((((.--....(((........))))))))).)))))..... ( -22.20) >DroSim_CAF1 51245 92 - 1 CCCUGCCAAAUGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCUGUAUUUUUUUUUUUUGCAUUUGUCAGCACAAAUGACUGCCUUUUG ..(((.(((((((((.....((((((((...............)))))))).........))))))))).)))................... ( -19.10) >DroEre_CAF1 55456 86 - 1 CCCUGCCAAACGCAAUAUUUGUGCAGCUUAUUUAUUUUAUUGUGGCUGCAUUU------UUUGCAUUUGUCAGCACAACUGACUGCCUUUUG ....((.....((((.....((((((((...............))))))))..------.))))....(((((.....))))).))...... ( -21.46) >DroYak_CAF1 50822 86 - 1 CCCUGCCAAACGCAAUAUUUGUGCAGUUUAUUUAUCUUAUUGUGGCUGCAUUU------UUUGCAUUUGUCAGCUCAAAUGACUGCCUUUUG ..(((.((((.((((.....((((((((...............))))))))..------.)))).)))).)))................... ( -18.36) >DroAna_CAF1 53733 85 - 1 CCCUGCCAAAGGCAAUAUUUAUGUCGCAUAUUUAUUUUAUUGGAGCUGCAUA-------CUUGCAUUUGUCAGCAUAACUGACUGCAAUUUG ...((((...)))).....(((((.((...((((......)))))).)))))-------.(((((...(((((.....)))))))))).... ( -17.10) >consensus CCCUGCCAAACGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCUGCAUUU______UUUGCAUUUGUCAGCACAAAUGACUGCCUUUUG ..(((.((((.((((.....((((((((...............)))))))).........)))).)))).)))................... (-15.01 = -14.73 + -0.27)
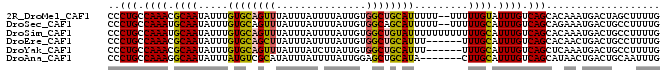
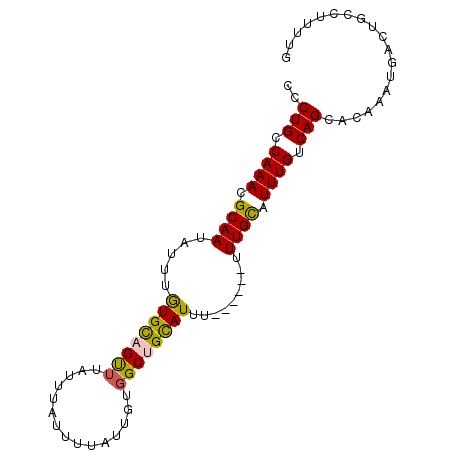
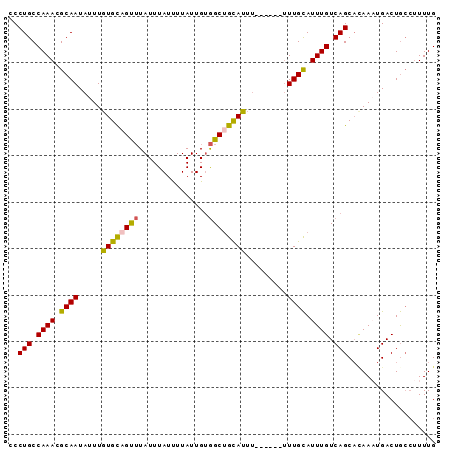
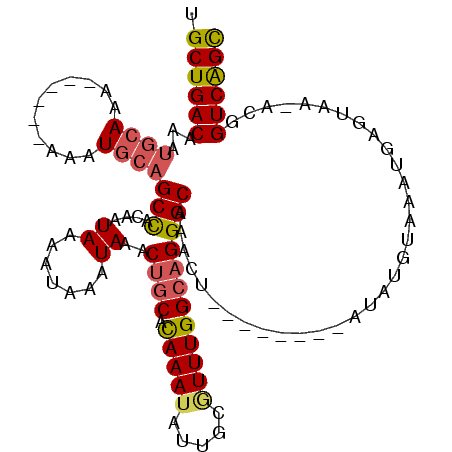
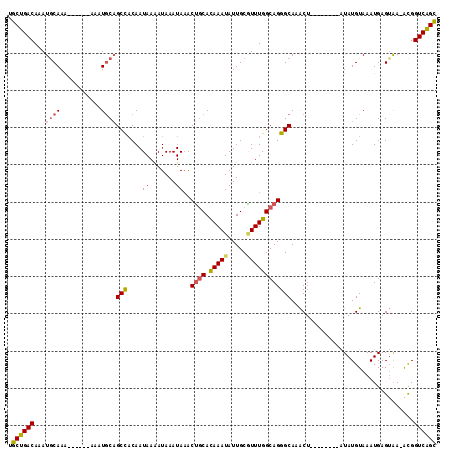
| Location | 10,221,784 – 10,221,896 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.04 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -16.49 |
| Energy contribution | -16.77 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10221784 112 + 20766785 UGCUGACAAAUACAAAAA--AAAAAUGCAGCCACAAUAAAAUAAAUAAACUGCACAAAUAUUGCGUUUGGCAGGGCAAACUACGUACAUAUAUGUAAAUGAGUAA-ACGGUCAGC .((((((...(((.....--.........(((....((.......))..((((.(((((.....))))))))))))....((((((....)))))).....))).-...)))))) ( -24.10) >DroSec_CAF1 50982 108 + 1 UGCUGACAAAUGCAAAAA--AAAAAUGCUGCCACAAUAAAAUAAAUAAACUGCACAAAUAUUGCAUUUGGCAGGGCAAACUGCGU----AUAUGUAAAUGAGUAA-ACGGUCAGC .((((((....(((....--.....)))((((....((.......))..((((.(((((.....))))))))))))).....(((----.(((........))).-))))))))) ( -23.00) >DroSim_CAF1 51262 110 + 1 UGCUGACAAAUGCAAAAAAAAAAAAUACAGCCACAAUAAAAUAAAUAAACUGCACAAAUAUUGCAUUUGGCAGGGCAAACUACGU----AUAUGUAAAUGAGUAA-ACGGUCAGC .((((((......................(((....((.......))..((((.(((((.....))))))))))))......(((----.(((........))).-))))))))) ( -22.50) >DroEre_CAF1 55473 101 + 1 UGCUGACAAAUGCAAA------AAAUGCAGCCACAAUAAAAUAAAUAAGCUGCACAAAUAUUGCGUUUGGCAGGGCAAACU--------AUAUGUAAAUGAGUAAAACGGUCAGC .((((((((((((((.------...((((((.................))))))......)))))))).(((.((....))--------...)))..............)))))) ( -23.83) >DroYak_CAF1 50839 101 + 1 AGCUGACAAAUGCAAA------AAAUGCAGCCACAAUAAGAUAAAUAAACUGCACAAAUAUUGCGUUUGGCAGGGCAAACU--------AUAUGUAAAUGAGUGAAGCGGUCGGC .((((((...((((..------...))))(((((...............((((.(((((.....))))))))).(((....--------...)))......)))..)).)))))) ( -24.30) >DroAna_CAF1 53750 100 + 1 UGCUGACAAAUGCAAG-------UAUGCAGCUCCAAUAAAAUAAAUAUGCGACAUAAAUAUUGCCUUUGGCAGGGCAAGCU--------GUAUGUAAAUGAGAGAAAUGGUCAGU .((((((...((((..-------..)))).((((((((.......)))...(((((....((((((......))))))...--------.)))))...)).))).....)))))) ( -24.50) >consensus UGCUGACAAAUGCAAA______AAAUGCAGCCACAAUAAAAUAAAUAAACUGCACAAAUAUUGCGUUUGGCAGGGCAAACU________AUAUGUAAAUGAGUAA_ACGGUCAGC .((((((...((((...........))))(((....((.......))..((((.(((((.....)))))))))))).................................)))))) (-16.49 = -16.77 + 0.28)

| Location | 10,221,784 – 10,221,896 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
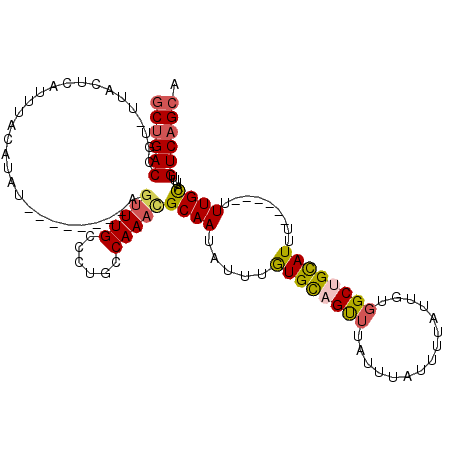
| Mean pairwise identity | 85.04 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -18.94 |
| Energy contribution | -19.17 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851375 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10221784 112 - 20766785 GCUGACCGU-UUACUCAUUUACAUAUAUGUACGUAGUUUGCCCUGCCAAACGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCUGCAUUUUU--UUUUUGUAUUUGUCAGCA ((((((.((-..(((.((.((((....)))).)))))..))......(((..(((.....((((((((...............))))))))....--...)))..))))))))). ( -25.26) >DroSec_CAF1 50982 108 - 1 GCUGACCGU-UUACUCAUUUACAUAU----ACGCAGUUUGCCCUGCCAAAUGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCAGCAUUUUU--UUUUUGCAUUUGUCAGCA ((((((...-................----..((((......)))).((((((((.....((((.(((...............))).))))....--...)))))))))))))). ( -25.26) >DroSim_CAF1 51262 110 - 1 GCUGACCGU-UUACUCAUUUACAUAU----ACGUAGUUUGCCCUGCCAAAUGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCUGUAUUUUUUUUUUUUGCAUUUGUCAGCA ((((((...-................----..((((......)))).((((((((.....((((((((...............)))))))).........)))))))))))))). ( -25.40) >DroEre_CAF1 55473 101 - 1 GCUGACCGUUUUACUCAUUUACAUAU--------AGUUUGCCCUGCCAAACGCAAUAUUUGUGCAGCUUAUUUAUUUUAUUGUGGCUGCAUUU------UUUGCAUUUGUCAGCA ((((((....................--------.(((((......)))))((((.....((((((((...............))))))))..------.))))....)))))). ( -28.96) >DroYak_CAF1 50839 101 - 1 GCCGACCGCUUCACUCAUUUACAUAU--------AGUUUGCCCUGCCAAACGCAAUAUUUGUGCAGUUUAUUUAUCUUAUUGUGGCUGCAUUU------UUUGCAUUUGUCAGCU ((.(((....................--------.(((((......)))))((((.....((((((((...............))))))))..------.))))....))).)). ( -21.36) >DroAna_CAF1 53750 100 - 1 ACUGACCAUUUCUCUCAUUUACAUAC--------AGCUUGCCCUGCCAAAGGCAAUAUUUAUGUCGCAUAUUUAUUUUAUUGGAGCUGCAUA-------CUUGCAUUUGUCAGCA .(((((....................--------((((((((........)))((((..((((...))))..))))......)))))(((..-------..)))....))))).. ( -19.20) >consensus GCUGACCGU_UUACUCAUUUACAUAU________AGUUUGCCCUGCCAAACGCAAUAUUUGUGCAGUUUAUUUAUUUUAUUGUGGCUGCAUUU______UUUGCAUUUGUCAGCA ((((((.............................(((((......)))))((((.....((((((((...............)))))))).........))))....)))))). (-18.94 = -19.17 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:06 2006