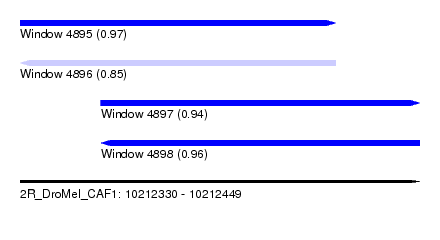
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,212,330 – 10,212,449 |
| Length | 119 |
| Max. P | 0.974405 |

| Location | 10,212,330 – 10,212,424 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -15.37 |
| Energy contribution | -15.15 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974405 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
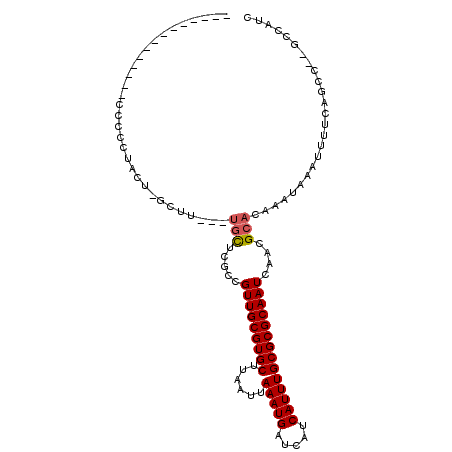
>2R_DroMel_CAF1 10212330 94 + 20766785 G-GAAACGGAGAA-AACCCAGAAAGAGAUGGC--GGCUGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACG (-....)((....-...))...........((--((((......((((((((((((.....))))))))))))((((......))))))).))).... ( -24.60) >DroGri_CAF1 47054 90 + 1 --------AGUUUAGGCCGAGUUAGGGAUAGCUAGACCGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACA --------.(((((((.(.(((((....))))).).))((....((((((((((((.....))))))))))))...)).......)))))........ ( -24.50) >DroEre_CAF1 45080 89 + 1 ------CGGGGAA-AACCCAGAAAGAGAUGGC--GGCUGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACG ------.(((...-..)))...........((--((((......((((((((((((.....))))))))))))((((......))))))).))).... ( -27.60) >DroYak_CAF1 41515 95 + 1 GGAAA-CGGGGAAAAGCCCAGAAAGAGAUGGC--GGCUGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACG (....-)(((......)))...........((--((((......((((((((((((.....))))))))))))((((......))))))).))).... ( -28.30) >DroAna_CAF1 45567 71 + 1 -------------------------AACUGGC--GGCUGGAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACG -------------------------.....((--((((.((...((((((((((((.....))))))))))))...)).........))).))).... ( -22.90) >DroPer_CAF1 23820 89 + 1 GGAGGCAGGA----GGCACUGGCA---AAGGA--GAAUGGAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACG ...(.(((..----....))).).---.....--((((((....((((((((((((.....))))))))))))...))))))................ ( -23.90) >consensus ______CGGGGAA_AGCCCAGAAAGAGAUGGC__GGCUGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACG ......................................((....((((((((((((.....))))))))))))...)).................... (-15.37 = -15.15 + -0.22)


| Location | 10,212,330 – 10,212,424 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.79 |
| Mean single sequence MFE | -16.71 |
| Consensus MFE | -13.98 |
| Energy contribution | -13.98 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10212330 94 - 20766785 CGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCAGCC--GCCAUCUCUUUCUGGGUU-UUCUCCGUUUC-C .(((((((((......(((((.....))))))))))))))........................--.(((........)))...-...........-. ( -15.60) >DroGri_CAF1 47054 90 - 1 UGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCGGUCUAGCUAUCCCUAACUCGGCCUAAACU-------- .(((((((((......(((((.....))))))))))))))....................((((.((.((....)).)).))))......-------- ( -18.60) >DroEre_CAF1 45080 89 - 1 CGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCAGCC--GCCAUCUCUUUCUGGGUU-UUCCCCG------ .(((((((((......(((((.....))))))))))))))........................--.............(((..-...))).------ ( -17.70) >DroYak_CAF1 41515 95 - 1 CGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCAGCC--GCCAUCUCUUUCUGGGCUUUUCCCCG-UUUCC .(((((((((......(((((.....))))))))))))))........................--.............(((......))).-..... ( -17.60) >DroAna_CAF1 45567 71 - 1 CGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUCCAGCC--GCCAGUU------------------------- .(((((((((......(((((.....))))))))))))))........................--.......------------------------- ( -14.00) >DroPer_CAF1 23820 89 - 1 CGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUCCAUUC--UCCUU---UGCCAGUGCC----UCCUGCCUCC .(((((((((......(((((.....))))))))))))))....((((................--.....---.....)))).----.......... ( -16.77) >consensus CGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCAGCC__GCCAUCUCUUUCUGGGCU_UUCCCCG______ .(((((((((......(((((.....)))))))))))))).......................................................... (-13.98 = -13.98 + -0.00)
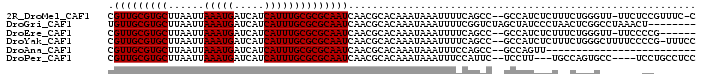
| Location | 10,212,354 – 10,212,449 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -28.45 |
| Consensus MFE | -16.08 |
| Energy contribution | -15.67 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.940160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10212354 95 + 20766785 GAUGGC--GGCUGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACGAAGAGCA--CAACU-AGUCGGGGG------UU---UU- ....((--((((......((((((((((((.....))))))))))))((((......))))))).)))..(((..((..--...))-..)))....------..---..- ( -25.90) >DroVir_CAF1 53581 95 + 1 GAUAGCUAGACCGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACGGCGCACA----AGCCAGAAACAGGACA----------- ..........((.........(((((((((((.(((((.(((((((.....)))))......)).))))))))))))))----)).........))...----------- ( -27.87) >DroGri_CAF1 47072 95 + 1 GAUAGCUAGACCGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACAGCGAACA----GGCAGGCAGGCGGCCA----------- ....(((.(...((....((((((((((((.....))))))))))))...)).............(((....)))..).----))).(((.....))).----------- ( -26.70) >DroEre_CAF1 45099 90 + 1 GAUGGC--GGCUGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACGAAGAGCA--CAACU-AGUAGGGG--------------- ....((--((((......((((((((((((.....))))))))))))((((......))))))).)))...........--.....-........--------------- ( -22.20) >DroAna_CAF1 45568 104 + 1 ACUGGC--GGCUGGAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACGGCGAGCA--GAAG--AGCUGGGAGCCUGGGUUCGGUUU .(..((--..((.((...((((((((((((.....))))))))))))...))..........((.(((....))).)).--..))--.))..)((((....))))..... ( -34.10) >DroPer_CAF1 23840 96 + 1 -AAGGA--GAAUGGAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACGGCGAAUCCGGAAAC-GGUAGCUGG------AC---CU- -.(((.--((((((....((((((((((((.....))))))))))))...))))))..............((((....(((....)-))..)))).------.)---))- ( -33.90) >consensus GAUGGC__GACUGAAAAUUUAUUUGUGCGUUGAUUGCGCGCAAAUGAUGAUCAUUUAAUUAAGCACGCAACGGCGAACA___AAGC_AGUAGGGGG______________ ........................((.(((((.(((((.(((((((.....)))))......)).)))))))))).))................................ (-16.08 = -15.67 + -0.42)
| Location | 10,212,354 – 10,212,449 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.30 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -14.27 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10212354 95 - 20766785 -AA---AA------CCCCCGACU-AGUUG--UGCUCUUCGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCAGCC--GCCAUC -..---..------.........-..(((--(((..((.(((((((((......(((((.....)))))))))))))).)).))))))..............--...... ( -21.80) >DroVir_CAF1 53581 95 - 1 -----------UGUCCUGUUUCUGGCU----UGUGCGCCGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCGGUCUAGCUAUC -----------......(((...((((----((((((..(((((((((......(((((.....))))))))))))))...))))))...........)))).))).... ( -25.70) >DroGri_CAF1 47072 95 - 1 -----------UGGCCGCCUGCCUGCC----UGUUCGCUGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCGGUCUAGCUAUC -----------.(((((..........----(((.((.((((((((((......(((((.....))))))))))))).)).)).)))..........)))))........ ( -24.65) >DroEre_CAF1 45099 90 - 1 ---------------CCCCUACU-AGUUG--UGCUCUUCGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCAGCC--GCCAUC ---------------........-..(((--(((..((.(((((((((......(((((.....)))))))))))))).)).))))))..............--...... ( -21.80) >DroAna_CAF1 45568 104 - 1 AAACCGAACCCAGGCUCCCAGCU--CUUC--UGCUCGCCGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUCCAGCC--GCCAGU ............((((...(((.--....--.))).((.(((((((((......(((((.....))))))))))))))....))..............))))--...... ( -23.50) >DroPer_CAF1 23840 96 - 1 -AG---GU------CCAGCUACC-GUUUCCGGAUUCGCCGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUCCAUUC--UCCUU- -.(---(.------.......((-(....)))....((.(((((((((......(((((.....))))))))))))))....))............))....--.....- ( -19.50) >consensus ______________CCCCCUACU_GCUU___UGCUCGCCGUUGCGUGCUUAAUUAAAUGAUCAUCAUUUGCGCGCAAUCAACGCACAAAUAAAUUUUCAGCC__GCCAUC ...............................(((.....(((((((((......(((((.....))))))))))))))....)))......................... (-14.27 = -14.35 + 0.08)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:04 2006