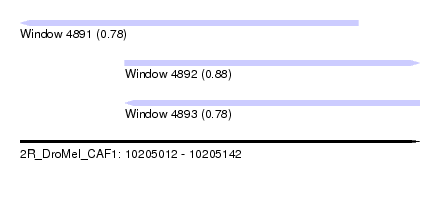
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,205,012 – 10,205,142 |
| Length | 130 |
| Max. P | 0.877587 |

| Location | 10,205,012 – 10,205,122 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 80.30 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -17.24 |
| Energy contribution | -19.80 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.781903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10205012 110 - 20766785 AAG-AUUGGAGGAU-AAUACCCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCAGUCUAACAAGCAGUCUAAAUAGGUA-UUAGGGA-UACCAGUAAAAUCAAUACUUG---- ...-...((.((..-....))))((((((((((......(((.((((((((((....)).))))))))....))).....((((-((...))-)))).......))))))))))---- ( -28.50) >DroSec_CAF1 34587 99 - 1 AAG-AUUGGGGGAU-AAUACCCCCAGGUGUUGAAUUCAGGGAAUU-----------GCCGCCUAACAAGCAGUCUAAAUAGGCA-UUAGGGA-UACCAGUAAAAUCAAUACUUG---- ...-...(((((..-....)))))(((((((((.....((.....-----------.))(((((...((....))...))))).-...((..-..)).......))))))))).---- ( -28.30) >DroSim_CAF1 34508 110 - 1 AAG-AUUGGGGGAU-AAUACCCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGCCUAACAAGCAGUCUAAAUAGGCA-UUAGGGA-UACCAGUAAAAUCAAUACUUG---- ...-...(((((..-....)))))(((((((((..........((((((((((.....))))))))))((.((((....)))).-...((..-..)).))....))))))))).---- ( -37.70) >DroEre_CAF1 37686 115 - 1 AAGAUUUGGGGAAU-AAUGCUCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGUCUAACAAGCAGUCUAAAUAGGCA-UUAGGAA-UACCAGUAAAAUCAAUACUUGGCUG .......((((...-....))))((((((((((......((..(((((((..(.....)..)))))))...((((....)))).-.......-..)).......)))))))))).... ( -27.72) >DroYak_CAF1 34053 112 - 1 AAGGAUUGGGGGAU-AAUACCCCUAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGUCUAACAAGCAGUCUAAAUAGGCA-UUAGGGAUUACCAGUAAAAUCAAUACUUG---- ......((((((..-....))))))((((((((..........(((((((..(.....)..)))))))((.((((....)))).-...((.....)).))....))))))))..---- ( -30.30) >DroAna_CAF1 39160 93 - 1 --------ACGGAUAAAUGUCCUUAGGUGUUGUAUUCACAGAAUUG---------UGCAACUUAACAAGCAGGCUAAACAGUCAAUGAGGGC-UUCCUGCAAAAUCAUU---GG---- --------..(((.....(((((((...(((((((..........)---------))))))..........((((....))))..)))))))-.)))............---..---- ( -19.90) >consensus AAG_AUUGGGGGAU_AAUACCCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGCCUAACAAGCAGUCUAAAUAGGCA_UUAGGGA_UACCAGUAAAAUCAAUACUUG____ .......(((((.......)))))(((((((((..........(((((((((.......)))))))))((.((((....)))).....((.....)).))....)))))))))..... (-17.24 = -19.80 + 2.56)

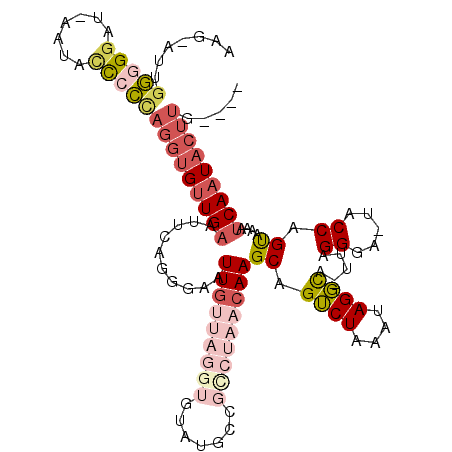
| Location | 10,205,046 – 10,205,142 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -18.84 |
| Consensus MFE | -13.60 |
| Energy contribution | -13.32 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877587 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10205046 96 + 20766785 UUUAGACUGCUUGUUAGACUGCAUACACCUAACAAUUCCCUGAAUUCAACACCUGGGGGUAUUAUCCUCCAAU-CUUUGCCCCUCUUAACAAUUUUC (((((.....(((((((...........)))))))....)))))..........(((((((............-...)))))))............. ( -15.66) >DroSec_CAF1 34621 85 + 1 UUUAGACUGCUUGUUAGGCGGC-----------AAUUCCCUGAAUUCAACACCUGGGGGUAUUAUCCCCCAAU-CUUUGUCCCUCUUAACAAUUUUC ..........((((((((.(((-----------((.....((....)).....((((((......))))))..-..)))))...))))))))..... ( -21.10) >DroSim_CAF1 34542 96 + 1 UUUAGACUGCUUGUUAGGCGGCAUACACCUAACAAUUCCCUGAAUUCAACACCUGGGGGUAUUAUCCCCCAAU-CUUUGCCCCUCUUAACAAUUUUC (((((.....((((((((.(.....).))))))))....))))).........((((((......))))))..-....................... ( -23.60) >DroEre_CAF1 37724 97 + 1 UUUAGACUGCUUGUUAGACGGCAUACACCUAACAAUUCCCUGAAUUCAACACCUGGGAGCAUUAUUCCCCAAAUCUUUGCCCCUCUUGGCAAUUUUC .......((((((((((..(.....)..))))))).((((..............)))))))...............(((((......)))))..... ( -17.94) >DroYak_CAF1 34088 97 + 1 UUUAGACUGCUUGUUAGACGGCAUACACCUAACAAUUCCCUGAAUUCAACACCUAGGGGUAUUAUCCCCCAAUCCUUGCCCCCUCUUAACAAUUUUC ..........((((((((.((((..........(((((...))))).........((((......)))).......))))...)).))))))..... ( -15.90) >consensus UUUAGACUGCUUGUUAGACGGCAUACACCUAACAAUUCCCUGAAUUCAACACCUGGGGGUAUUAUCCCCCAAU_CUUUGCCCCUCUUAACAAUUUUC ..........((((((((.((............(((((...))))).......((((((......)))))).........)).)).))))))..... (-13.60 = -13.32 + -0.28)

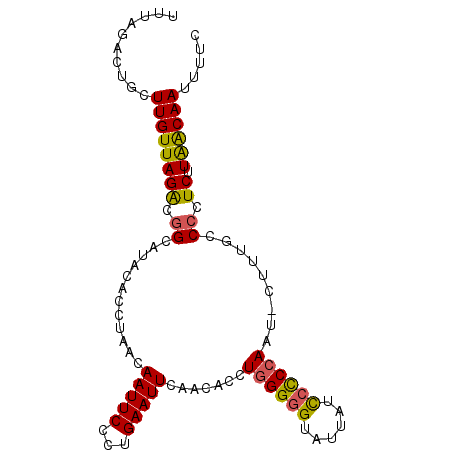
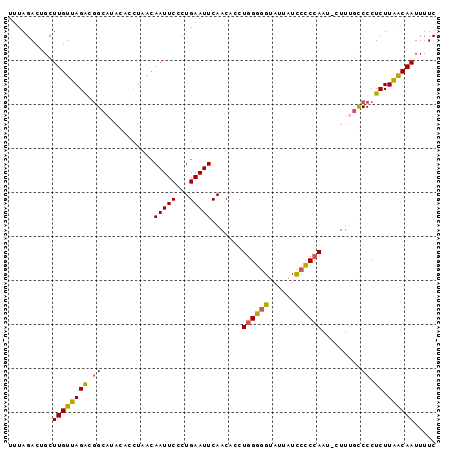
| Location | 10,205,046 – 10,205,142 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 89.14 |
| Mean single sequence MFE | -26.20 |
| Consensus MFE | -18.04 |
| Energy contribution | -20.04 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10205046 96 - 20766785 GAAAAUUGUUAAGAGGGGCAAAG-AUUGGAGGAUAAUACCCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCAGUCUAACAAGCAGUCUAAA .....(((((......)))))((-(((((((..(((((((....)))))))..)))......((((((((((....)).)))))))).))))))... ( -24.10) >DroSec_CAF1 34621 85 - 1 GAAAAUUGUUAAGAGGGACAAAG-AUUGGGGGAUAAUACCCCCAGGUGUUGAAUUCAGGGAAUU-----------GCCGCCUAACAAGCAGUCUAAA ((...((((((.(.((((((...-.(((((((......))))))).)))).(((((...)))))-----------.)).).))))))....)).... ( -23.80) >DroSim_CAF1 34542 96 - 1 GAAAAUUGUUAAGAGGGGCAAAG-AUUGGGGGAUAAUACCCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGCCUAACAAGCAGUCUAAA ....((((((..(((...(((..-.(((((((......)))))))...)))..))).......(((((((((.....)))))))))))))))..... ( -32.20) >DroEre_CAF1 37724 97 - 1 GAAAAUUGCCAAGAGGGGCAAAGAUUUGGGGAAUAAUGCUCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGUCUAACAAGCAGUCUAAA (((...((((......))))((.((((((((........)))))))).))...)))..(((.(((((((..(.....)..)))))))....)))... ( -25.60) >DroYak_CAF1 34088 97 - 1 GAAAAUUGUUAAGAGGGGGCAAGGAUUGGGGGAUAAUACCCCUAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGUCUAACAAGCAGUCUAAA ....((((((....(((..(((.(.(((((((......))))))).).)))..))).......((((((..(.....)..))))))))))))..... ( -25.30) >consensus GAAAAUUGUUAAGAGGGGCAAAG_AUUGGGGGAUAAUACCCCCAGGUGUUGAAUUCAGGGAAUUGUUAGGUGUAUGCCGUCUAACAAGCAGUCUAAA ....(((((...(((...(((....(((((((......)))))))...)))..)))......((((((((((.....)))))))))))))))..... (-18.04 = -20.04 + 2.00)

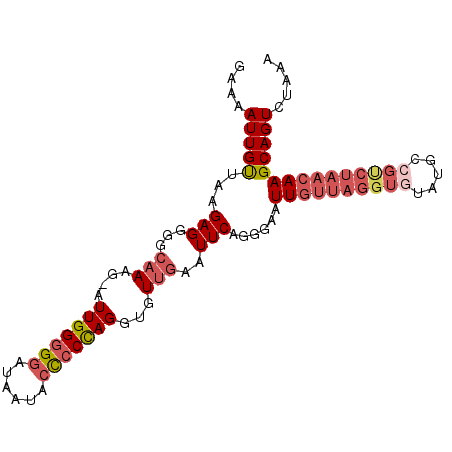

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:44:00 2006