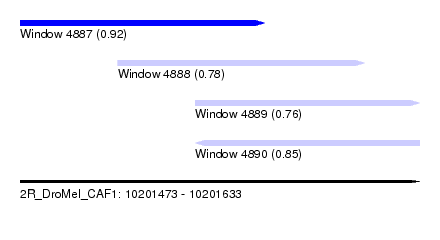
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,201,473 – 10,201,633 |
| Length | 160 |
| Max. P | 0.915584 |

| Location | 10,201,473 – 10,201,571 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 74.58 |
| Mean single sequence MFE | -13.77 |
| Consensus MFE | -6.63 |
| Energy contribution | -6.80 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10201473 98 + 20766785 AAAUAGAAUGGCACAUG-UACAUACAUACAUAUGUAUAUGUUUAAACCAAACG---CAACGGUCAAUCAAUUACAGAAUGGCCACAGAAUAAGCAUAAUAAA ..........(((((((-((((((......)))))))))))............---....(((((.((.......)).))))).........))........ ( -20.50) >DroSec_CAF1 31179 75 + 1 AAAUAGAAUG--------UACA----------------UAUAUAAACCAAACG---CAACGGUCAAUCAAUUACAGAAUGGCCACAGAAUAAGCAUAAUAAA ..........--------....----------------...............---....(((((.((.......)).)))))................... ( -7.70) >DroSim_CAF1 30983 87 + 1 AAAUAGAAUG--------UACAUAUGUACAUACA----UAUAUAAACCAAACG---CAACGGUCAAUCAAUUACAGAAUGGCCACAGAAUAAGCAUAAUAAA .....(.(((--------(((....)))))).).----...............---....(((((.((.......)).)))))................... ( -13.60) >DroEre_CAF1 34158 94 + 1 AAAUAGAAUGGCACAUG-UACCUACAUACAUGUA----UGCAUAAAGCGAACG---CAGCGGUCAAUCAAUUACAGAAUGGCCACAGAAUCAGCAUAAUAAA ............(((((-((......)))))))(----(((.....((....)---).(.(((((.((.......)).))))).).......))))...... ( -20.20) >DroYak_CAF1 30456 87 + 1 AAACAGAAUGGCAUAUA-UA----CACACAUAUC----UACAUAUACCAAACG---CAUCGGUCAAUC---UACAGAAUGGCCGCAGAAUAAGCAUAAUAAA ........(((.((((.-((----..........----)).)))).)))....---....(((((.((---....)).)))))((.......))........ ( -10.90) >DroAna_CAF1 31573 89 + 1 AAA-AGAA---CACAUUAUAC-AACACACAU--A----AACAUUAAG--AGUGUAAUAUCGGUCAAUCAAUUACGAAAUGGCCGCAGAAUAAGCAUAAUAAA ...-....---...(((((.(-...((((..--.----.........--.)))).....((((((.((......))..))))))........).)))))... ( -9.72) >consensus AAAUAGAAUG_CACAU__UACAUACAUACAUAUA____UACAUAAACCAAACG___CAACGGUCAAUCAAUUACAGAAUGGCCACAGAAUAAGCAUAAUAAA ............................................................(((((.((.......)).)))))................... ( -6.63 = -6.80 + 0.17)

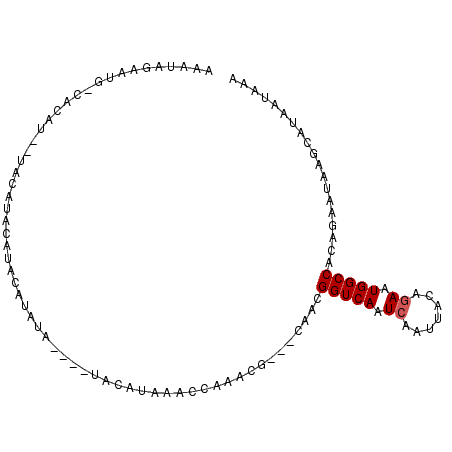
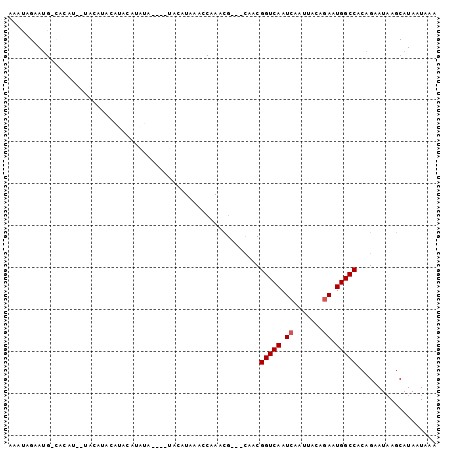
| Location | 10,201,512 – 10,201,611 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.09 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -11.88 |
| Energy contribution | -12.52 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10201512 99 + 20766785 UUUAAAC--C---AAACG---CA-ACGGUCAAUCAAUUACAGAAUGGCCACAGAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUCCCGA ...((((--(---(...(---(.-..(((((.((.......)).)))))((((..(((((((.................)))))))..))))))...))))))..... ( -21.63) >DroPse_CAF1 13339 103 + 1 UGGAGAC--UCUGGAGAG---CUUCUGUUCAAUCAAUUACCGAAUGGCCACAAAAUAGGCAUAAUAAAUUCGCAAUUAAAUGCUUAAAUUGUGCAAAUGGCUGUCCAA .(....)--..((((.((---((..(((.((((........((((.(((........))).......))))(((......)))....)))).)))...)))).)))). ( -24.20) >DroEre_CAF1 34193 99 + 1 CAUAAAG--C---GAACG---CA-GCGGUCAAUCAAUUACAGAAUGGCCACAGAAUCAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGCUUCCCGA ......(--(---....)---)(-(((((((.((.......)).))))).........(((((((......(((......)))....))))))).....)))...... ( -20.60) >DroWil_CAF1 33380 98 + 1 CAUAAACACUGUG---------A-ACAGUCAAUCAAUUACAAAAUGGCCACAAAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUGCCAA .......((((..---------.-.))))...............(((((((((..(((((((.................)))))))..)))))..........)))). ( -17.43) >DroAna_CAF1 31602 100 + 1 CAUUAAG-------AGUGUAAUA-UCGGUCAAUCAAUUACGAAAUGGCCGCAGAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUACCAA .......-------.(..((((.-.((((((.((......))..)))))).....(((((((.................))))))).))))..)...(((....))). ( -18.43) >DroPer_CAF1 13177 103 + 1 UGGAGGC--UCUGGAGAG---CUUCUGUUCAAUCAAUUACCGAAUGCCCACAAAAUAGGCAUAAUAAAUUCGUAAUUAAAUGCUUAAAUUGUGCAAAUGGCUGUCCAA .((((((--((....)))---)))))...............((..((((((((..(((((((.................)))))))..))))).....)))..))... ( -24.53) >consensus CAUAAAC__U___GAGAG___CA_ACGGUCAAUCAAUUACAGAAUGGCCACAAAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGCUUCCCAA ..........................(((((.((.......)).))))).........(((((((......(((......)))....)))))))...(((....))). (-11.88 = -12.52 + 0.64)

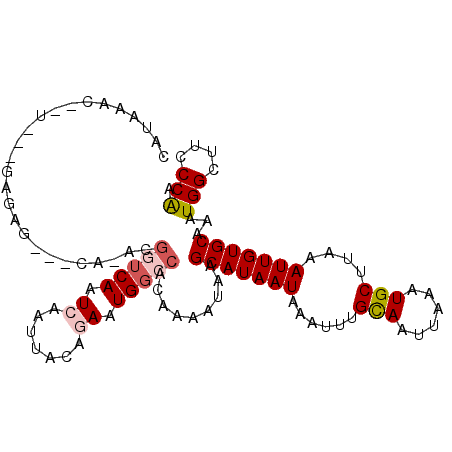
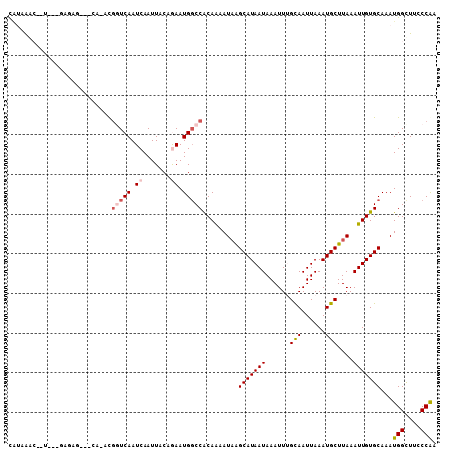
| Location | 10,201,543 – 10,201,633 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -19.76 |
| Consensus MFE | -13.63 |
| Energy contribution | -13.05 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.758139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10201543 90 + 20766785 AGAAUGGCCACAGAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUCCCGAUAUGGUGAC-------GCGGUUUGAGUUC .((((...(((((..(((((((.................)))))))..)))))(((((.((.((((.....)).)).-------)).))))).)))) ( -19.13) >DroPse_CAF1 13374 90 + 1 CGAAUGGCCACAAAAUAGGCAUAAUAAAUUCGCAAUUAAAUGCUUAAAUUGUGCAAAUGGCUGUCCAAUAUGGCGACCGCCGCGGCGGUC------- ......((((........(((((((......(((......)))....)))))))...(((....)))...))))((((((....))))))------- ( -26.20) >DroSim_CAF1 31042 90 + 1 AGAAUGGCCACAGAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUCCCGAUAUGGUGAC-------GCGGUUUGAGUUC .((((...(((((..(((((((.................)))))))..)))))(((((.((.((((.....)).)).-------)).))))).)))) ( -19.13) >DroWil_CAF1 33410 81 + 1 AAAAUGGCCACAAAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUGCCAAUAUCGGAAGAG-----AG----------- ....(((((((((..(((((((.................)))))))..)))))..........))))...((....)).-----..----------- ( -15.93) >DroYak_CAF1 30515 90 + 1 AGAAUGGCCGCAGAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGCUUCCCGAUAUGGUGAC-------GCGGUUCGAGCAC .....((((((.......(((((((......(((......)))....))))))).................(....)-------))))))....... ( -19.50) >DroAna_CAF1 31634 79 + 1 GAAAUGGCCGCAGAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUACCAAUCCGGCGGG-------GG----------- .......((((.......(((((((......(((......)))....)))))))...(((....))).....)))).-------..----------- ( -18.70) >consensus AGAAUGGCCACAGAAUAAGCAUAAUAAAUUUGCAAUUAAAUGCUUAAAUUGUGCAAAUGGUUUCCCAAUAUGGUGAC_______GCGGUU_______ ....(.((((........(((((((......(((......)))....)))))))...(((....)))...)))).)..................... (-13.63 = -13.05 + -0.58)

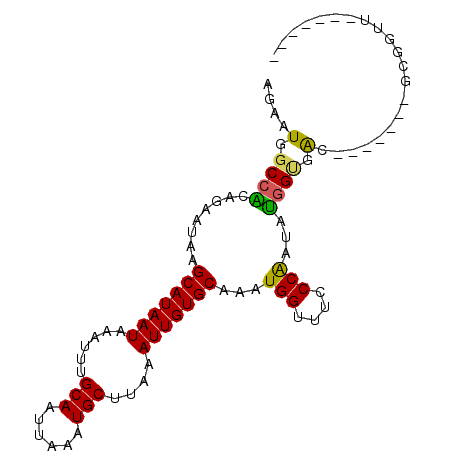
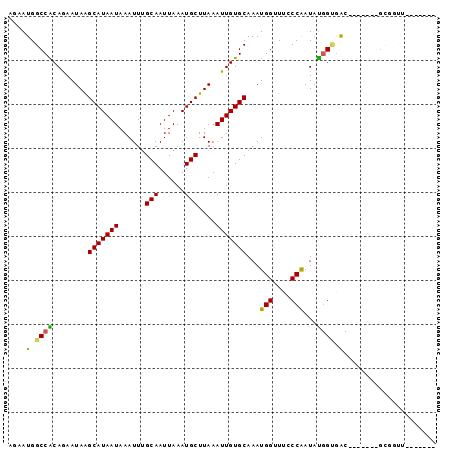
| Location | 10,201,543 – 10,201,633 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 79.53 |
| Mean single sequence MFE | -18.95 |
| Consensus MFE | -11.22 |
| Energy contribution | -11.72 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10201543 90 - 20766785 GAACUCAAACCGC-------GUCACCAUAUCGGGAAACCAUUUGCACAAUUUAAGCAUUUAAUUGCAAAUUUAUUAUGCUUAUUCUGUGGCCAUUCU ...........((-------...........((....)).....((((...(((((((.(((........)))..)))))))...))))))...... ( -17.80) >DroPse_CAF1 13374 90 - 1 -------GACCGCCGCGGCGGUCGCCAUAUUGGACAGCCAUUUGCACAAUUUAAGCAUUUAAUUGCGAAUUUAUUAUGCCUAUUUUGUGGCCAUUCG -------((((((....))))))((((((.(((....)))...(((.(((....(((......)))......))).)))......))))))...... ( -27.50) >DroSim_CAF1 31042 90 - 1 GAACUCAAACCGC-------GUCACCAUAUCGGGAAACCAUUUGCACAAUUUAAGCAUUUAAUUGCAAAUUUAUUAUGCUUAUUCUGUGGCCAUUCU ...........((-------...........((....)).....((((...(((((((.(((........)))..)))))))...))))))...... ( -17.80) >DroWil_CAF1 33410 81 - 1 -----------CU-----CUCUUCCGAUAUUGGCAAACCAUUUGCACAAUUUAAGCAUUUAAUUGCAAAUUUAUUAUGCUUAUUUUGUGGCCAUUUU -----------..-----............((((....(....)(((((..(((((((.(((........)))..)))))))..))))))))).... ( -17.20) >DroYak_CAF1 30515 90 - 1 GUGCUCGAACCGC-------GUCACCAUAUCGGGAAGCCAUUUGCACAAUUUAAGCAUUUAAUUGCAAAUUUAUUAUGCUUAUUCUGCGGCCAUUCU ......(((((((-------...........((....))............(((((((.(((........)))..)))))))....))))...))). ( -16.10) >DroAna_CAF1 31634 79 - 1 -----------CC-------CCCGCCGGAUUGGUAAACCAUUUGCACAAUUUAAGCAUUUAAUUGCAAAUUUAUUAUGCUUAUUCUGCGGCCAUUUC -----------..-------...(((((((((((((.....)))).)))))(((((((.(((........)))..))))))).....))))...... ( -17.30) >consensus _______AACCGC_______GUCACCAUAUCGGGAAACCAUUUGCACAAUUUAAGCAUUUAAUUGCAAAUUUAUUAUGCUUAUUCUGUGGCCAUUCU ...............................((....)).....((((...(((((((.(((........)))..)))))))...))))........ (-11.22 = -11.72 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:57 2006