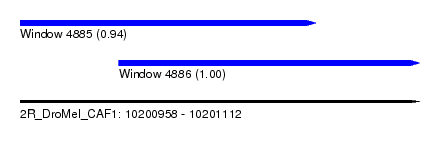
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,200,958 – 10,201,112 |
| Length | 154 |
| Max. P | 0.999510 |

| Location | 10,200,958 – 10,201,072 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.43 |
| Mean single sequence MFE | -34.09 |
| Consensus MFE | -31.25 |
| Energy contribution | -30.85 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.937809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10200958 114 + 20766785 UGGAUUGGGGAAAAAUGUGGCCGAAAAUGGUGCGAGGGCGUCUGUUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAGCGGAGAAAUCG ..((((.............(((......)))((..(..(((((((((((((((((((((((.((.....))..)))))).))))))))))))).)))).)..)).....)))). ( -32.40) >DroSec_CAF1 29102 114 + 1 UGGAUUGGGGAAAAAUGUGGCCGAAAAUGGUGCGAGGGCGUCUGCUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAACGGAGAAAUCG ..((((.........(((.((((.....(..((....))..)(((((((((((((((((((.((.....))..)))))).))))))))))))))))).)))........)))). ( -34.73) >DroSim_CAF1 29365 114 + 1 UGGAUUGGGGAAAAAUGUGGCCGAAAAUGGUGCGAGGGCGUCUGCUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAACGGAGAAAUCG ..((((.........(((.((((.....(..((....))..)(((((((((((((((((((.((.....))..)))))).))))))))))))))))).)))........)))). ( -34.73) >DroEre_CAF1 33650 112 + 1 UGGAUUGGGGAAAAA-GUGGCC-AAAAUGGUGCGAGGGCGUCUGCAGGCGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCUGCAUGGCGACGAGCGGAGAAAUCG .........(..(((-((((((-(((((....((((..((((....))))...))).)....)))).))))))))))..).(((((..((((.((....)).))))..))))). ( -34.40) >DroYak_CAF1 29904 112 + 1 UGGAUUGGGGAAAAA-GUGACCGAAAAUGGUGCGAGGGCGUCUGCAGGAGAGUUUCUGAAAAGU-UCUGGCCAUUUUAGCAGAUUUUCCUGCAUGGCGACGAGCUGAGAAAUCG ....((((.......-....)))).......((..(..(((((((((((((((((((((((.((-....))..)))))).))))))))))))).)))).)..)).((....)). ( -34.20) >consensus UGGAUUGGGGAAAAAUGUGGCCGAAAAUGGUGCGAGGGCGUCUGCUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAGCGGAGAAAUCG ..((((..........((.(((......(..((....))..)(((((((((((((((((((.((.....))..)))))).))))))))))))).))).)).........)))). (-31.25 = -30.85 + -0.40)

| Location | 10,200,996 – 10,201,112 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.97 |
| Mean single sequence MFE | -37.18 |
| Consensus MFE | -32.40 |
| Energy contribution | -32.28 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.87 |
| SVM decision value | 3.67 |
| SVM RNA-class probability | 0.999510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10200996 116 + 20766785 CGUCUGUUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAGCGGAGAAAUCGAGCUAAUAAAUUGACUCGGGGAAAUUGUGUCAAACAUGCC (((((((((((((((((((((((.((.....))..)))))).))))))))))))).))))...(((.((....))..)))......(((((.(((.....))).)))))....... ( -34.90) >DroSec_CAF1 29140 116 + 1 CGUCUGCUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAACGGAGAAAUCGAGCUAAUAAAUUGACUCGGGAAAACUGUGUCAAAUAUGCC (((((((((((((((((((((((.((.....))..)))))).))))))))))))).)))).......((....))...........(((((.(((.....))).)))))....... ( -36.80) >DroSim_CAF1 29403 116 + 1 CGUCUGCUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAACGGAGAAAUCGAGCUAAUAAAUUGACUCGGGAAAACUGUGUCAAAUAUGCC (((((((((((((((((((((((.((.....))..)))))).))))))))))))).)))).......((....))...........(((((.(((.....))).)))))....... ( -36.80) >DroEre_CAF1 33686 116 + 1 CGUCUGCAGGCGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCUGCAUGGCGACGAGCGGAGAAAUCGAACUAAUAAAUUGACUCUGGGAAACUGUGUCAAAUACGCC ((((((((((.((((((((((((.((.....))..)))))).)))))).)))))).))))....(((((....))...........(((((.(.((....))).)))))...))). ( -35.90) >DroYak_CAF1 29941 115 + 1 CGUCUGCAGGAGAGUUUCUGAAAAGU-UCUGGCCAUUUUAGCAGAUUUUCCUGCAUGGCGACGAGCUGAGAAAUCGAGCUAAUAAAUUGACUCUAGGAAACUGUGUCAAAUAUGCC (((((((((((((((((((((((.((-....))..)))))).))))))))))))).))))...((((((....)).))))......(((((.(.((....))).)))))....... ( -41.50) >consensus CGUCUGCUGGAGAAUUUCUGAAAAGUUUCUGGCCAUUUUAGCAGAUUUUCCAGCAUGGCGACGAGCGGAGAAAUCGAGCUAAUAAAUUGACUCGGGGAAACUGUGUCAAAUAUGCC (((((((((((((((((((((((.((.....))..)))))).))))))))))))).)))).(((.........)))..........(((((.(((.....))).)))))....... (-32.40 = -32.28 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:54 2006