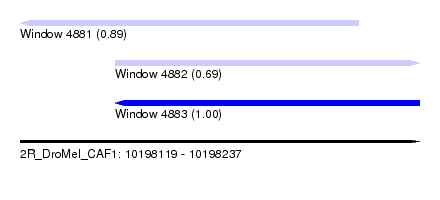
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,198,119 – 10,198,237 |
| Length | 118 |
| Max. P | 0.996149 |

| Location | 10,198,119 – 10,198,219 |
|---|---|
| Length | 100 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.39 |
| Mean single sequence MFE | -19.36 |
| Consensus MFE | -18.23 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894545 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10198119 100 - 20766785 GCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUACCACAUUCGCACAAACGCACA--CACAUAAACAAAGCAACCACUUUUCUCGCCAUGCAAA (((((((...)))))))......((((((......(((...........)))......((...--............))............))))))..... ( -18.36) >DroEre_CAF1 30953 99 - 1 GCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUGCCACACUCGCACAAACGCACA--CACAUAAACA-AGCGGCCACUUUUCUCGCCAUGCAAA (((..(((((((((........)))))).)))...((((.(((.......)))...))))...--..........-)))(((..........)))....... ( -21.40) >DroYak_CAF1 26853 102 - 1 GCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUACCACACUCGCACAAACGCACACACACAUAAACAAAGCAGCCACUUUUCUCGCCAUGCAAA (((((((...)))))))......((((((......((((.................))))..............((((......))))...))))))..... ( -18.33) >consensus GCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUACCACACUCGCACAAACGCACA__CACAUAAACAAAGCAGCCACUUUUCUCGCCAUGCAAA (((((((...)))))))......((((((......(((...........)))......((.................))............))))))..... (-18.23 = -18.23 + 0.00)

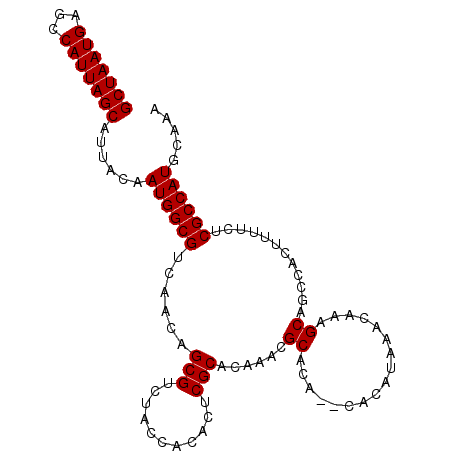
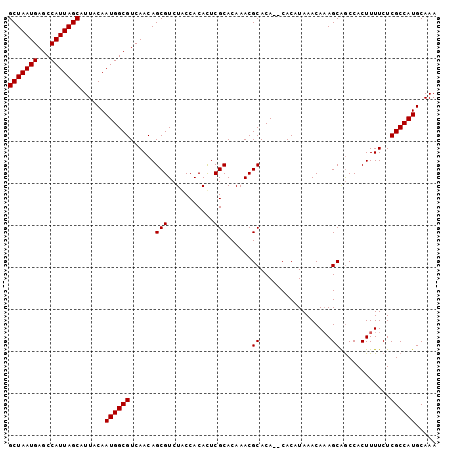
| Location | 10,198,147 – 10,198,237 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -20.81 |
| Energy contribution | -21.25 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685672 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10198147 90 + 20766785 GUUUAUGUG--UGUGCGUUUGUGCGAAUGUGGUAGACGCUGUUGACGCCAUUGUAAUGCUAAUGGCUCAUUAGCACUUUGAUUUUUUCGCAC ....((((.--...))))..(((((((.....((((.((((.(((.(((((((......)))))))))).))))..)))).....))))))) ( -25.70) >DroEre_CAF1 30980 90 + 1 GUUUAUGUG--UGUGCGUUUGUGCGAGUGUGGCAGACGCUGUUGACGCCAUUGUAAUGCUAAUGGCUCAUUAGCACUUUGAUUUUUUCGCAC ....((((.--...))))..(((((((.....((((.((((.(((.(((((((......)))))))))).))))..)))).....))))))) ( -27.20) >DroYak_CAF1 26881 92 + 1 GUUUAUGUGUGUGUGCGUUUGUGCGAGUGUGGUAGACGCUGUUGACGCCAUUGUAAUGCUAAUGGCUCAUUAGCACUUUGAUUUUUUCGCAC ....((((......))))..(((((((.....((((.((((.(((.(((((((......)))))))))).))))..)))).....))))))) ( -25.60) >DroAna_CAF1 25265 78 + 1 -------------UGUCUGUGUGAGAGUGUGGUGGAUCCUGUUGACGCCAUUGUAAUGCUAAUGGCUCAUUAGCACUUUGAUUUUUUCGCU- -------------.....(((.((((((..((((....(((.(((.(((((((......)))))))))).)))))))...)))))).))).- ( -19.90) >consensus GUUUAUGUG__UGUGCGUUUGUGCGAGUGUGGUAGACGCUGUUGACGCCAUUGUAAUGCUAAUGGCUCAUUAGCACUUUGAUUUUUUCGCAC ....................(((((((.....((((.((((.(((.(((((((......)))))))))).))))..)))).....))))))) (-20.81 = -21.25 + 0.44)


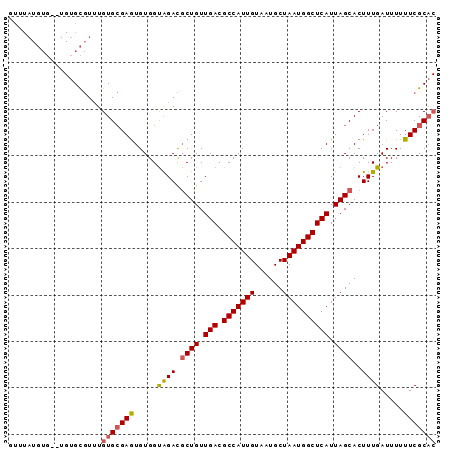
| Location | 10,198,147 – 10,198,237 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 86.81 |
| Mean single sequence MFE | -21.97 |
| Consensus MFE | -18.25 |
| Energy contribution | -19.50 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.996149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10198147 90 - 20766785 GUGCGAAAAAAUCAAAGUGCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUACCACAUUCGCACAAACGCACA--CACAUAAAC (((((((........((((((((((...)))))))))).....(((((.....))))).......))))))).........--......... ( -25.60) >DroEre_CAF1 30980 90 - 1 GUGCGAAAAAAUCAAAGUGCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUGCCACACUCGCACAAACGCACA--CACAUAAAC ((((((.........((((((((((...)))))))))).....(((((.....)))))........)))))).........--......... ( -24.50) >DroYak_CAF1 26881 92 - 1 GUGCGAAAAAAUCAAAGUGCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUACCACACUCGCACAAACGCACACACACAUAAAC ((((((.........((((((((((...)))))))))).....(((((.....)))))........)))))).................... ( -24.50) >DroAna_CAF1 25265 78 - 1 -AGCGAAAAAAUCAAAGUGCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGGAUCCACCACACUCUCACACAGACA------------- -.(.((.........((((((((((...))))))))))....(((.(((.....))))))........)).).......------------- ( -13.30) >consensus GUGCGAAAAAAUCAAAGUGCUAAUGAGCCAUUAGCAUUACAAUGGCGUCAACAGCGUCUACCACACUCGCACAAACGCACA__CACAUAAAC ((((((.........((((((((((...)))))))))).....(((((.....)))))........)))))).................... (-18.25 = -19.50 + 1.25)

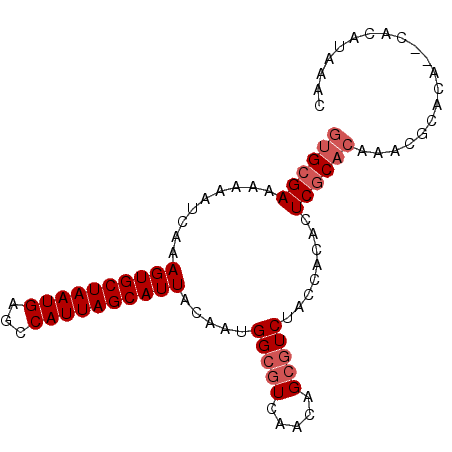
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:51 2006