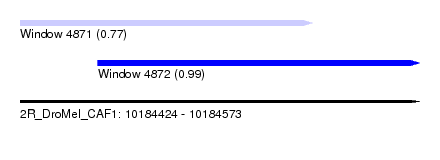
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,184,424 – 10,184,573 |
| Length | 149 |
| Max. P | 0.988834 |

| Location | 10,184,424 – 10,184,533 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.80 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -23.79 |
| Energy contribution | -22.85 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771701 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10184424 109 + 20766785 GAAUGAC----GACUCU-UG-----UCAUCCACAGAUCUAUAUGGUGAUACCACCGGACGGCGGAUUCGGUUGGGUAAUCAUGGUGCUGUCCUUCCUCGCCCAGCUGAUAAUCGAUGGU ..(((((----((...)-))-----))))(((..(((.....(((((....)))))..........((((((((((......((......))......))))))))))..)))..))). ( -31.00) >DroSim_CAF1 12635 109 + 1 GAAUGAC----GACUCU-UG-----UCAUCCACAGAUCUAUAUGGUGAUACCGCCGGACGGCGGAUUCGGCUGGGUAAUCAUGGUGUUGUCCUUCCUCGCCCAGCUGAUAGUCGAUGGU ......(----((((..-((-----(((.(((.(......).))))))))(((((....)))))..((((((((((......((......))......)))))))))).)))))..... ( -40.40) >DroEre_CAF1 17279 104 + 1 AAAUGAC-------U---UG-----UCAUCCGCAGAUCUAUAUGGUGAUACCACCGGACGGCGGAUUCGGCUGGGUAAUCAUGGUGCUGUCCUUCCUCGCCCAGCUGAUAAUCGAUGGC .......-------(---..-----((((((((...(((....((((....)))))))..))))))((((((((((......((......))......)))))))))).....))..). ( -36.50) >DroWil_CAF1 15735 112 + 1 UAAUUAUUUGCCUUUCU-UG-----U-AUUGUUAGAUCUACAUGGUGAUACCACCAGAUGGUGGCUUUGGUUGGGUCAUCUUAAUCCUGAGUUUCCUAGCCCAACUAAUUGUCGAUGGU .........(((..((.-..-----.-..........(((..(((((....)))))..))).(((.((((((((((...((((....)))).......))))))))))..))))).))) ( -27.70) >DroYak_CAF1 12955 106 + 1 GAAUGAC-------UCG-UG-----UCAUUCACAGAUCUACAUGGUGAUACCACCGGACGGCGGAUUUGGCUGGGUCAUCAUGGUGCUGUCCUUUCUCGCCCAGCUAAUAAUCGAUGGU (((((((-------...-.)-----)))))).((((((..(.(((((....)))))....)..))))))(((((((......((......))......))))))).............. ( -37.80) >DroMoj_CAF1 5283 114 + 1 AACUGAC----AUUCGCUUGCAAUUACCU-CGUAGAUCUACAUGGCGAUCCCGCCGGACGGUGGCUUUGGCUGGGUCGUGCUCAUUCUCAGCUUUCUUGCCCAGCUCAUUGUGGAUGGC ......(----((((((..((...(((((-((((....))).(((((....))))))).))))))...((((((((...(((.......)))......))))))))....))))))).. ( -36.00) >consensus GAAUGAC_______UCU_UG_____UCAUCCACAGAUCUACAUGGUGAUACCACCGGACGGCGGAUUCGGCUGGGUAAUCAUGGUGCUGUCCUUCCUCGCCCAGCUGAUAAUCGAUGGU ..........................................(((((....)))))..((.(((..((((((((((......((......))......))))))))))...))).)).. (-23.79 = -22.85 + -0.94)

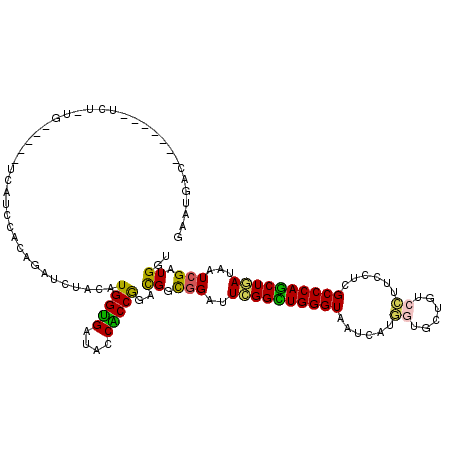
| Location | 10,184,453 – 10,184,573 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.67 |
| Mean single sequence MFE | -41.17 |
| Consensus MFE | -25.75 |
| Energy contribution | -26.07 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10184453 120 + 20766785 AUAUGGUGAUACCACCGGACGGCGGAUUCGGUUGGGUAAUCAUGGUGCUGUCCUUCCUCGCCCAGCUGAUAAUCGAUGGUUUAAUUUUUACCAUUGGGGUCUUAUUGCCAUCCAUUGCCC ....((((....))))(((.(((((..((((((((((......((......))......))))))))))...((((((((.........)))))))).......))))).)))....... ( -41.50) >DroEre_CAF1 17303 120 + 1 AUAUGGUGAUACCACCGGACGGCGGAUUCGGCUGGGUAAUCAUGGUGCUGUCCUUCCUCGCCCAGCUGAUAAUCGAUGGCCUUAUUUUUACCAUUGGAAUCCUAUUGCCCUCCAUUGCCA ....((((....))))(((.(((((((((((((((((......((......))......))))))))......((((((...........))))))))))))....))).)))....... ( -43.20) >DroWil_CAF1 15767 120 + 1 ACAUGGUGAUACCACCAGAUGGUGGCUUUGGUUGGGUCAUCUUAAUCCUGAGUUUCCUAGCCCAACUAAUUGUCGAUGGUAUUAUAUUCUCAAUUGGCAUUCUCUUGCCCUACAUGAGCA .(((((((((((((((....)))(((.((((((((((...((((....)))).......))))))))))..)))...))))))))..........((((......))))...)))).... ( -37.70) >DroYak_CAF1 12981 120 + 1 ACAUGGUGAUACCACCGGACGGCGGAUUUGGCUGGGUCAUCAUGGUGCUGUCCUUUCUCGCCCAGCUAAUAAUCGAUGGUCUUAUAUUUACCAUCGGAGUCUUGUUGCCAUCCAUUGCCA ....((((....))))(((.(((((..((((((((((......((......))......))))))))))...((((((((.........)))))))).......))))).)))....... ( -43.20) >DroMoj_CAF1 5317 120 + 1 ACAUGGCGAUCCCGCCGGACGGUGGCUUUGGCUGGGUCGUGCUCAUUCUCAGCUUUCUUGCCCAGCUCAUUGUGGAUGGCAUCAUUUUCUCGAUUGGCAUUCUGCUGCCCGCCAUGGAGA .(((((((((((((((....))))((..(((((((((...(((.......)))......))))))).))..))))))((((((........))).(((.....))))))))))))).... ( -44.90) >DroAna_CAF1 12070 120 + 1 ACAUGAUUAUACCACCGGACGGUGGCUUCGGCUGGUGGAUCAUGGUCCUCAGCUUCUUUUCCCAGGUGAUCGUUGACGGCAUCAUAUUCUCCAUUGGCAUCCUGCUGCCGGCAAUAAGCA .((((((....(((((((.(((.....))).))))))))))))).......((((...((.((.((((.(((....)))))))............((((......)))))).)).)))). ( -36.50) >consensus ACAUGGUGAUACCACCGGACGGCGGAUUCGGCUGGGUCAUCAUGGUGCUGACCUUCCUCGCCCAGCUGAUAAUCGAUGGCAUUAUAUUCACCAUUGGCAUCCUGUUGCCCUCCAUUGCCA ....((((....))))(((.(((((..((((((((((......(........)......))))))))))...(((((((...........))))))).......))))).)))....... (-25.75 = -26.07 + 0.31)
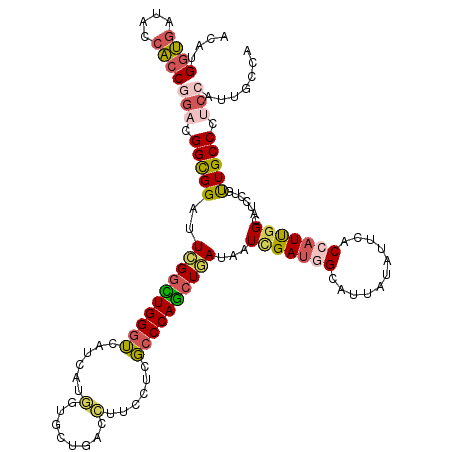
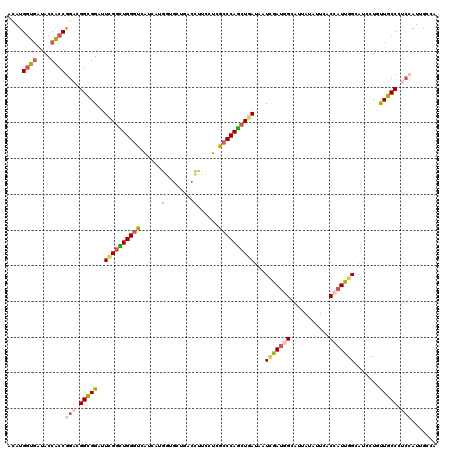
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:41 2006