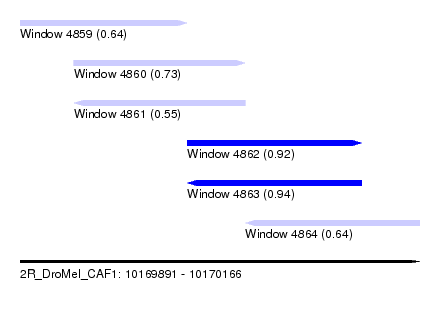
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,169,891 – 10,170,166 |
| Length | 275 |
| Max. P | 0.938609 |

| Location | 10,169,891 – 10,170,006 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.38 |
| Mean single sequence MFE | -24.19 |
| Consensus MFE | -19.37 |
| Energy contribution | -20.09 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642408 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10169891 115 + 20766785 ---CCACAUCCAGUGCAACAUAUUGACCAGCACUUGUUUGCUUUU--UUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUC ---...((..((((((.............)))).))..)).....--...............((((((((.((......((((.................)))).))))))))))..... ( -24.95) >DroSec_CAF1 9680 117 + 1 ---CCACAUCCAGUGCAACAUAUUGACCAGCACUUGUUUGCUUUUCUUUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUC ---...((..((((((.............)))).))..))......................((((((((.((......((((.................)))).))))))))))..... ( -24.95) >DroSim_CAF1 9499 117 + 1 ---CCACAUCCAGUGCAACAUAUUGACCAGCACUUGUUUGCUUUUCUUUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUC ---...((..((((((.............)))).))..))......................((((((((.((......((((.................)))).))))))))))..... ( -24.95) >DroEre_CAF1 9669 111 + 1 ---CCACAUCCAGUGCAACAUAUUGACCAGCACUUGUUUGCUU------UUUUAUUAUUGCUGUGGAGCAACCCUCCUUGCUAUCUAUACGUCCAAACUUUGGCUGGUGGUCCGCCCAUC ---((.....(((((.....)))))((((((....(((((...------..........((...((((.....))))..))............)))))....)))))))).......... ( -23.41) >DroYak_CAF1 10130 111 + 1 UUGCCACAUCCAGUGCAACAUAUUGGCCAGCACUUGUUUGCUU------UUUUAUUAUUGCUGUGGACCAACACUCCGUGCAAUCUAUAUGUCCAAUCUUUGGCUG---GUCCACCCAUC ((((.((.....)))))).....((((((((....(.(((..(------...(((.(((((.(((......))).....)))))..))).)..))).)....))))---).)))...... ( -22.70) >consensus ___CCACAUCCAGUGCAACAUAUUGACCAGCACUUGUUUGCUUUU__UUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUC ......((..((((((.............)))).))..))......................((((((((..((.....((((.................)))).))))))))))..... (-19.37 = -20.09 + 0.72)

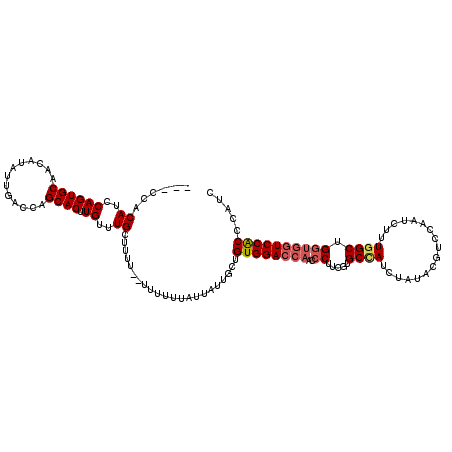
| Location | 10,169,928 – 10,170,046 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -19.43 |
| Energy contribution | -21.75 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10169928 118 + 20766785 CUUUU--UUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUCUGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAU .....--...........((((((((((((.((......((((.................)))).)))))))))).......(((((((((...........)))))))))....)))). ( -30.53) >DroSec_CAF1 9717 120 + 1 CUUUUCUUUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUCUGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAU ..................((((((((((((.((......((((.................)))).)))))))))).......(((((((((...........)))))))))....)))). ( -30.53) >DroSim_CAF1 9536 120 + 1 CUUUUCUUUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUCUGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAU ..................((((((((((((.((......((((.................)))).)))))))))).......(((((((((...........)))))))))....)))). ( -30.53) >DroEre_CAF1 9706 101 + 1 CUU------UUUUAUUAUUGCUGUGGAGCAACCCUCCUUGCUAUCUAUACGUCCAAACUUUGGCUGGUGGUCCGCCCAUCUGGGUAUAAAUUUAUGCACAUUAUUUG------------- ...------.........(((...(((((((......))))...((((.((.(((.....))).)))))))))((((....))))..........))).........------------- ( -17.30) >DroYak_CAF1 10170 111 + 1 CUU------UUUUAUUAUUGCUGUGGACCAACACUCCGUGCAAUCUAUAUGUCCAAUCUUUGGCUG---GUCCACCCAUCUGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAU ...------.........((((((((((((.(((...)))..........(.(((.....))))))---)))))).......(((((((((...........)))))))))....)))). ( -26.20) >consensus CUUUU__UUUUUUAUUAUUGCUGUGGACCAACCCUUCGUGCCAUCUAUACGUCCAAUCUUUGGCUGGUGGUCCACCCAUCUGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAU ..................((((((((((((..((.....((((.................)))).)))))))))).......(((((((((...........)))))))))....)))). (-19.43 = -21.75 + 2.32)

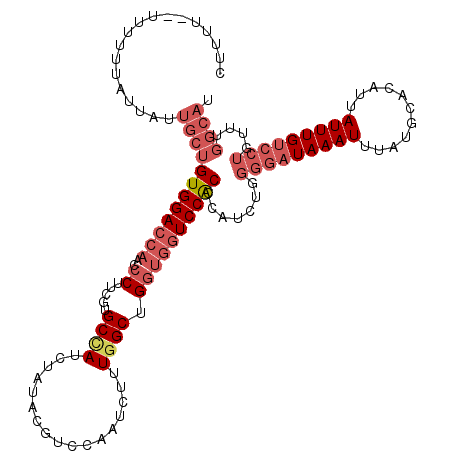
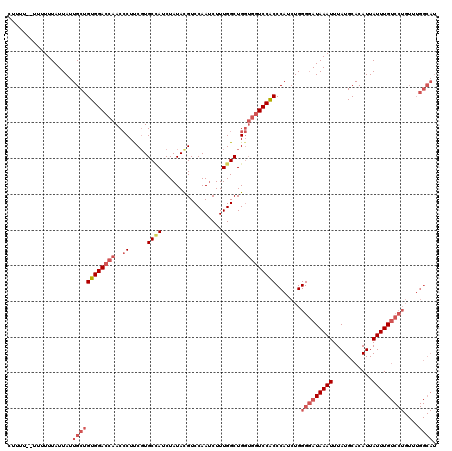
| Location | 10,169,928 – 10,170,046 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -18.62 |
| Energy contribution | -20.02 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10169928 118 - 20766785 AUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCAGAUGGGUGGACCACCAGCCAAAGAUUGGACGUAUAGAUGGCACGAAGGGUUGGUCCACAGCAAUAAUAAAAAA--AAAAG .((((.....)).)).......(((............((....))((((((((.(.((((...((.......))...)))).(....)).)))))))).)))...........--..... ( -25.90) >DroSec_CAF1 9717 120 - 1 AUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCAGAUGGGUGGACCACCAGCCAAAGAUUGGACGUAUAGAUGGCACGAAGGGUUGGUCCACAGCAAUAAUAAAAAAAGAAAAG .((((.....)).)).......(((............((....))((((((((.(.((((...((.......))...)))).(....)).)))))))).))).................. ( -25.90) >DroSim_CAF1 9536 120 - 1 AUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCAGAUGGGUGGACCACCAGCCAAAGAUUGGACGUAUAGAUGGCACGAAGGGUUGGUCCACAGCAAUAAUAAAAAAAGAAAAG .((((.....)).)).......(((............((....))((((((((.(.((((...((.......))...)))).(....)).)))))))).))).................. ( -25.90) >DroEre_CAF1 9706 101 - 1 -------------CAAAUAAUGUGCAUAAAUUUAUACCCAGAUGGGCGGACCACCAGCCAAAGUUUGGACGUAUAGAUAGCAAGGAGGGUUGCUCCACAGCAAUAAUAAAA------AAG -------------.........(((....((((((((((((((.(((((....)).)))...))))))..)))))))).....((((.....))))...))).........------... ( -27.00) >DroYak_CAF1 10170 111 - 1 AUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCAGAUGGGUGGAC---CAGCCAAAGAUUGGACAUAUAGAUUGCACGGAGUGUUGGUCCACAGCAAUAAUAAAA------AAG .(((......((((.((((.((((((...((((((((((.....)).)))(---(((.......)))).....)))))))))))...)))).))))...))).........------... ( -23.90) >consensus AUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCAGAUGGGUGGACCACCAGCCAAAGAUUGGACGUAUAGAUGGCACGAAGGGUUGGUCCACAGCAAUAAUAAAAAA__AAAAG ..........((((.(((..(((((....((((((..((((.(((.(((....))).)))....))))....)))))).)))))....))).))))........................ (-18.62 = -20.02 + 1.40)


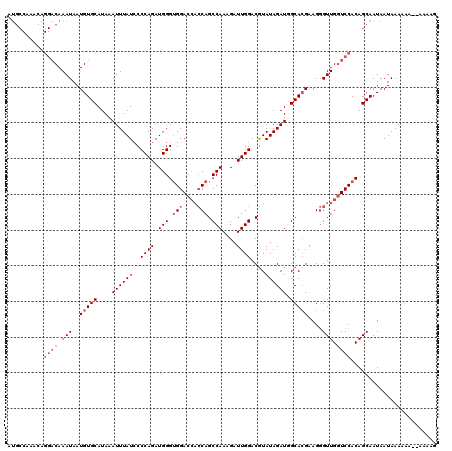
| Location | 10,170,006 – 10,170,126 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -31.60 |
| Energy contribution | -31.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10170006 120 + 20766785 UGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAUGUCGCGUUGAUUUUUGCGCAAUCCUCUUGCACAUUUCCAAAUGCCAUUUGUAUGCAAACUAACUGCGCUUUAUGCCUCCA .((((((((((...........)))))))).....((((((..((((.....(((((((((.....))))..........((((.....)))))))))......))))..)))))).)). ( -31.60) >DroSec_CAF1 9797 120 + 1 UGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAUGUCGCGUUGAUUUUUGCGCAAUCCUCUUGCACAUUUUCAAAUGCCAUUUGUAUGCAAACUAACUGCGCUUUAUGCCUCCA .((((((((((...........)))))))).....((((((..((((.....(((((((((.....))))..........((((.....)))))))))......))))..)))))).)). ( -31.60) >DroSim_CAF1 9616 120 + 1 UGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAUGUCGCGUUGAUUUUUGCGCAAUCCUCUUGCACAUUUCCAAAUGCCAUUUGUAUGCAAACUAACUGCGCUUUAUGCCUCCA .((((((((((...........)))))))).....((((((..((((.....(((((((((.....))))..........((((.....)))))))))......))))..)))))).)). ( -31.60) >DroYak_CAF1 10241 120 + 1 UGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAUGUCGCGUUGAUUUUUGCGCAAUCCUCUUGCACAUUUCCAAAUGCCAUUUGUAUGCAAACUAACUGCGCUUUAUGCCUCCA .((((((((((...........)))))))).....((((((..((((.....(((((((((.....))))..........((((.....)))))))))......))))..)))))).)). ( -31.60) >consensus UGGGGAUAAAUUUAUGCACAUUAUUUGUCCUGUUUGGCAUGUCGCGUUGAUUUUUGCGCAAUCCUCUUGCACAUUUCCAAAUGCCAUUUGUAUGCAAACUAACUGCGCUUUAUGCCUCCA .((((((((((...........)))))))).....((((((..((((.....(((((((((.....))))..........((((.....)))))))))......))))..)))))).)). (-31.60 = -31.60 + -0.00)

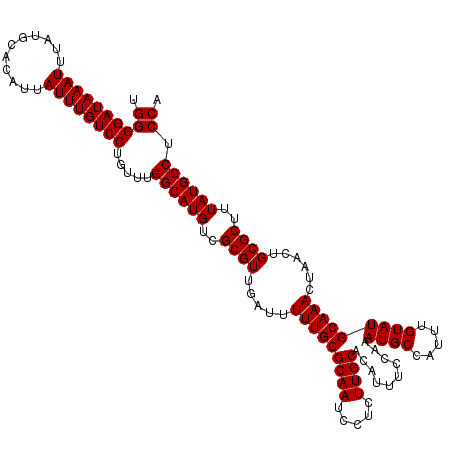
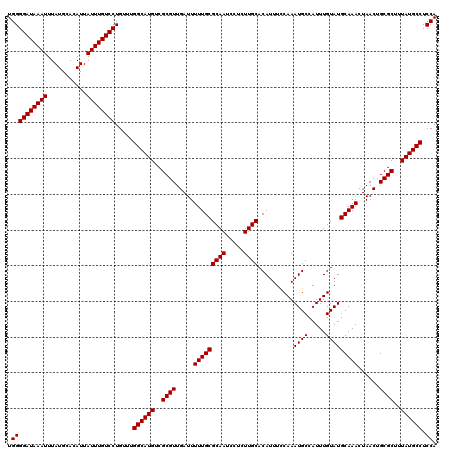
| Location | 10,170,006 – 10,170,126 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -33.30 |
| Energy contribution | -33.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.07 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.27 |
| SVM RNA-class probability | 0.938609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10170006 120 - 20766785 UGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGACAUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCA .((.((.(((((((((((((..(((((((((((((...)))))....))))))))..))))))))..........((.((((.((.....)).......)))))).....))))))))). ( -33.30) >DroSec_CAF1 9797 120 - 1 UGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGAAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGACAUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCA .((.((.(((((((((((((..(((((((((((((...)))))....))))))))..))))))))..........((.((((.((.....)).......)))))).....))))))))). ( -33.30) >DroSim_CAF1 9616 120 - 1 UGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGACAUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCA .((.((.(((((((((((((..(((((((((((((...)))))....))))))))..))))))))..........((.((((.((.....)).......)))))).....))))))))). ( -33.30) >DroYak_CAF1 10241 120 - 1 UGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGACAUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCA .((.((.(((((((((((((..(((((((((((((...)))))....))))))))..))))))))..........((.((((.((.....)).......)))))).....))))))))). ( -33.30) >consensus UGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGACAUGCCAAACAGGACAAAUAAUGUGCAUAAAUUUAUCCCCA .((.((.(((((((((((((..(((((((((((((...)))))....))))))))..))))))))..........((.((((.((.....)).......)))))).....))))))))). (-33.30 = -33.30 + 0.00)

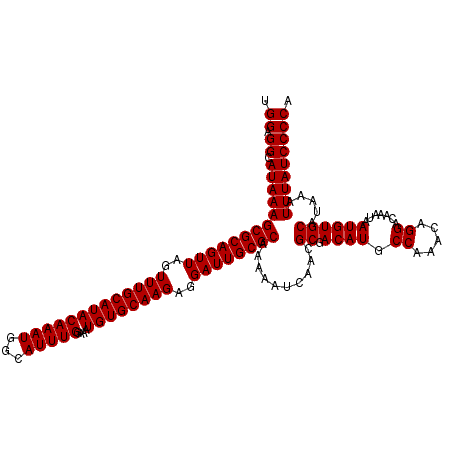
| Location | 10,170,046 – 10,170,166 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.75 |
| Mean single sequence MFE | -31.17 |
| Consensus MFE | -30.60 |
| Energy contribution | -30.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10170046 120 - 20766785 GAUUGGCAUGAGGAUUGAAUUGAGAAUGGCAUGAUGACCAUGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGAC ..............(((..((((..(((((.....).))))...........((((((((..(((((((((((((...)))))....))))))))..)))))))).....))))..))). ( -28.90) >DroSec_CAF1 9837 120 - 1 GAUUGGCAUGAGGAUUGAGUUGAGAAUGGCAUGAUGACCAUGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGAAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGAC ..............(((.(((((..(((((.....).))))...........((((((((..(((((((((((((...)))))....))))))))..)))))))).....))))).))). ( -31.70) >DroSim_CAF1 9656 120 - 1 GAUUGGCAUGAGGAUUGAGUUGAGAAUGGCAUGAUGACCAUGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGAC ..............(((.(((((..(((((.....).))))...........((((((((..(((((((((((((...)))))....))))))))..)))))))).....))))).))). ( -31.70) >DroYak_CAF1 10281 120 - 1 GAUUGGCAUCAGGAUUGAGUUGAGAAUGGCAUGAUGACCAUGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGAC .....((.((......))(((((..(((((.....).))))...........((((((((..(((((((((((((...)))))....))))))))..)))))))).....)))))))... ( -32.40) >consensus GAUUGGCAUGAGGAUUGAGUUGAGAAUGGCAUGAUGACCAUGGAGGCAUAAAGCGCAGUUAGUUUGCAUACAAAUGGCAUUUGGAAAUGUGCAAGAGGAUUGCGCAAAAAUCAACGCGAC ..............(((.(((((..(((((.....).))))...........((((((((..(((((((((((((...)))))....))))))))..)))))))).....))))).))). (-30.60 = -30.85 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:33 2006