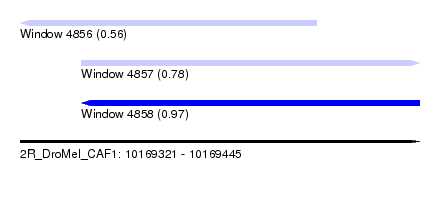
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,169,321 – 10,169,445 |
| Length | 124 |
| Max. P | 0.966025 |

| Location | 10,169,321 – 10,169,413 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 78.03 |
| Mean single sequence MFE | -16.10 |
| Consensus MFE | -9.55 |
| Energy contribution | -10.80 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10169321 92 - 20766785 UGGCAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAUUGUUGUUGUUAUCAUUCGUAUACACUCAUAUUU-------AUUAUAGCUAUUUUCUAUGU-UU ....((((((.......(((.(((.((((...)))).))).)))(((((.((..((((......))))..-------)).))))).......)))))-). ( -13.00) >DroSec_CAF1 9115 93 - 1 UGGAAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAAUGUGGUUUUAAUCAUUCGUAUUCACUCAUAUUU-------GUUAUAGCUAUUUCCUAUACCUU .(((((...........(((..(((((((...)))))))..)))..........((....((........-------))....))..)))))........ ( -15.90) >DroSim_CAF1 8934 93 - 1 UGGCAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAAUGUGGUUUUAAUCAUUCGUAUUCACUCAUAUUU-------GUUAUAGCUAUUUCCUAUACCUU ((((.............(((..(((((((...)))))))..)))................((........-------))....))))............. ( -15.00) >DroYak_CAF1 9522 91 - 1 UGGCAUCAUAUUUAAUUAAAUGCAUU-UCUUUGAGAAUGUGGUUUUAAUCAUCUGGGGUCAUGAAAAUGACAUCUUUUCGAUAGCCAUUUCC-------- (((((((((((((..(((((......-..))))))))))))))....(((....((((((((....))))).)))....))).)))).....-------- ( -20.50) >consensus UGGCAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAAUGUGGUUUUAAUCAUUCGUAUUCACUCAUAUUU_______GUUAUAGCUAUUUCCUAUAC_UU ((((.((..........((((((((((((...))))))))))))..........)).))))....................................... ( -9.55 = -10.80 + 1.25)

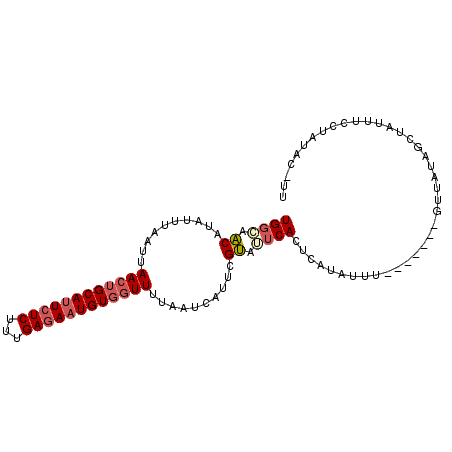
| Location | 10,169,340 – 10,169,445 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -14.40 |
| Consensus MFE | -3.54 |
| Energy contribution | -4.18 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.25 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10169340 105 + 20766785 AAU-------AAAUAUGAGUGUAUACGAAUGAUAACAACAACAAUCUCAAAGAGAAUGCAGUUAAUUAAAUAUGUUGCCACCAUAUCAAUGAAUACAA----CUCAUCA---CAAUCC-A ...-------....((((((((((.((..(((((....(((((.((((...)))).....(((.....))).)))))......))))).)).)))).)----)))))..---......-. ( -14.50) >DroSec_CAF1 9135 105 + 1 AAC-------AAAUAUGAGUGAAUACGAAUGAUUAAAACCACAUUCUCAAAGAGAAUGCAGUUAAUUAAAUAUGUUUCCAGCAUAUCAAUGAAUACAA----CUCAUCA---CAAUCC-A ...-------....((((((.........(((((((.....(((((((...)))))))...))))))).((((((.....))))))...........)----)))))..---......-. ( -18.10) >DroSim_CAF1 8954 105 + 1 AAC-------AAAUAUGAGUGAAUACGAAUGAUUAAAACCACAUUCUCAAAGAGAAUGCAGUUAAUUAAAUAUGUUGCCACCAUAUCAAUGAAUACAA----CUCAUCA---CAAUCC-A ...-------........((((.......(((((((.....(((((((...)))))))...))))))).....((((....(((....)))....)))----)...)))---).....-. ( -16.30) >DroEre_CAF1 9115 92 + 1 CGAAACG--GAAAUUUUAAUGACCACGGAUGAUUAAAACCACAUGCUCAAAGA-----------------UAUGAUGCCACCAUAUCAAUGAAUACAA----AUCAUCA---CAACCC-- .......--..................(((((((.................((-----------------((((.......))))))..((....)))----)))))).---......-- ( -11.30) >DroYak_CAF1 9534 119 + 1 CGAAAAGAUGUCAUUUUCAUGACCCCAGAUGAUUAAAACCACAUUCUCAAAGA-AAUGCAUUUAAUUAAAUAUGAUGCCACCAUGUUAAUGAAUACAACCCAAUCACCAAUUCAACUCCA ......(((.(((((..((((......................(((.....))-)..(((((...........)))))...))))..)))))..........)))............... ( -11.80) >consensus AAA_______AAAUAUGAGUGAAUACGAAUGAUUAAAACCACAUUCUCAAAGAGAAUGCAGUUAAUUAAAUAUGUUGCCACCAUAUCAAUGAAUACAA____CUCAUCA___CAAUCC_A ..........................((.((..........(((((((...)))))))...........(((((.......)))))..........................)).))... ( -3.54 = -4.18 + 0.64)

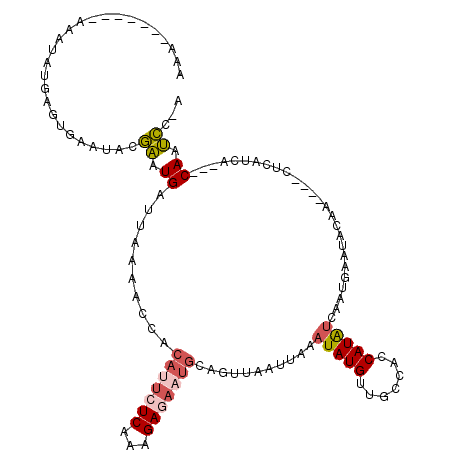
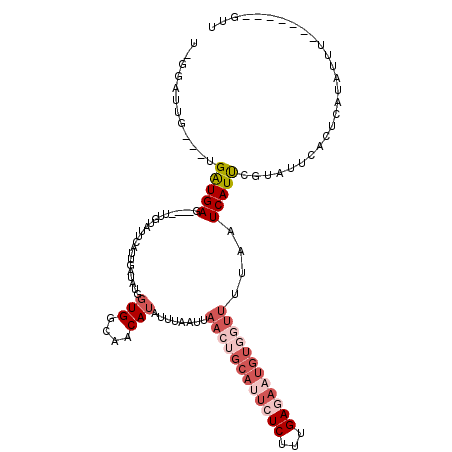
| Location | 10,169,340 – 10,169,445 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.20 |
| Mean single sequence MFE | -24.81 |
| Consensus MFE | -9.18 |
| Energy contribution | -11.78 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10169340 105 - 20766785 U-GGAUUG---UGAUGAG----UUGUAUUCAUUGAUAUGGUGGCAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAUUGUUGUUGUUAUCAUUCGUAUACACUCAUAUUU-------AUU .-......---..(((((----(.((((....(((.((((((((((((.............(((.((((...)))).)))))))))))))))))).))))))))))....-------... ( -26.81) >DroSec_CAF1 9135 105 - 1 U-GGAUUG---UGAUGAG----UUGUAUUCAUUGAUAUGCUGGAAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAAUGUGGUUUUAAUCAUUCGUAUUCACUCAUAUUU-------GUU .-.(((..---..(((((----(.((((....((((....((....)).........(((..(((((((...)))))))..)))...))))...))))..))))))....-------))) ( -24.30) >DroSim_CAF1 8954 105 - 1 U-GGAUUG---UGAUGAG----UUGUAUUCAUUGAUAUGGUGGCAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAAUGUGGUUUUAAUCAUUCGUAUUCACUCAUAUUU-------GUU (-((((..---.((((((----((((..((((....))))..)))))..........(((..(((((((...)))))))..)))....)))))...))))).........-------... ( -25.90) >DroEre_CAF1 9115 92 - 1 --GGGUUG---UGAUGAU----UUGUAUUCAUUGAUAUGGUGGCAUCAUA-----------------UCUUUGAGCAUGUGGUUUUAAUCAUCCGUGGUCAUUAAAAUUUC--CGUUUCG --(((...---(((((((----.....((((..((((((((...))))))-----------------))..))))((((.(((.......)))))))))))))).....))--)...... ( -21.30) >DroYak_CAF1 9534 119 - 1 UGGAGUUGAAUUGGUGAUUGGGUUGUAUUCAUUAACAUGGUGGCAUCAUAUUUAAUUAAAUGCAUU-UCUUUGAGAAUGUGGUUUUAAUCAUCUGGGGUCAUGAAAAUGACAUCUUUUCG ..(((..((...((((((((..(..(((((.((((...((..((((.............))))...-)).)))))))))..)..).)))))))....(((((....))))).))..))). ( -25.72) >consensus U_GGAUUG___UGAUGAG____UUGUAUUCAUUGAUAUGGUGGCAACAUAUUUAAUUAACUGCAUUCUCUUUGAGAAUGUGGUUUUAAUCAUUCGUAUUCACUCAUAUUU_______GUU ............(((((......................(((....)))........((((((((((((...))))))))))))....)))))........................... ( -9.18 = -11.78 + 2.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:27 2006