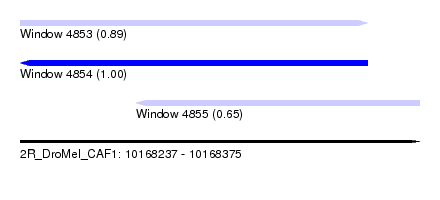
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,168,237 – 10,168,375 |
| Length | 138 |
| Max. P | 0.999435 |

| Location | 10,168,237 – 10,168,357 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -32.06 |
| Energy contribution | -32.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10168237 120 + 20766785 AAUAAUUUAGCAACCGGAGCGGACAAUAAAAUUUCGAGCCCAAAUGUCAUACGCAUGCAUUUGCAUAUGAUUUAGCCGGCGCUGGGUGAACAGUAUCCGUAUCCGAAUCGGAUUCGGAAU .............((((((((((..............(((.....((((((.(((......))).))))))......)))((((......)))).))))).((((...)))))))))... ( -33.80) >DroSec_CAF1 8103 120 + 1 AAUAAUUUAGCAACCGGAGCGGACAAUAAAAUUUCGAGCCCAAAUGUCAUACGCAUGCAUUUGCAUAUGAUUUAGCCGGCGCUGGGUGAACAGUAUCCGUAUCCGAAUCGGAUUCGGAAU .............((((((((((..............(((.....((((((.(((......))).))))))......)))((((......)))).))))).((((...)))))))))... ( -33.80) >DroSim_CAF1 7943 120 + 1 AAUAAUUUAGCAACCGGAGCGGACAAUAAAAUUUCGAGCCCAAAUGUCAUACGCAUGCAUUUGCAUAUGAUUUAGCCGGCGCUGGGUGAACAGUAUCCGUAUCCGAAUCGGAUUCGGAAU .............((((((((((..............(((.....((((((.(((......))).))))))......)))((((......)))).))))).((((...)))))))))... ( -33.80) >DroEre_CAF1 8101 120 + 1 AAUAAUUUAGCAACCGGAGCGGACAAUAAAAUUUCGAGCCCAAAUGUCAUACGCAUGCAUUUGCAUAUGAUUUAGCCGGCGCUGGGUGAACAGUAUCCGUAUCCGAAUCGGAUUCGGAAU .............((((((((((..............(((.....((((((.(((......))).))))))......)))((((......)))).))))).((((...)))))))))... ( -33.80) >DroYak_CAF1 7885 120 + 1 AAUAAUUUAGCAACCGGAGCGGACAAUAAAAUUUCGAGCCCAAAUGUCAUACGCAUGCAUUUGCAUAUGAUUUAGCCGGCGCUGGGUGAACAGUAUCCGUAUCCAAAUCGGAUUCAGAAU ...............((((((((..............(((.....((((((.(((......))).))))))......)))((((......)))).))))).)))................ ( -28.90) >consensus AAUAAUUUAGCAACCGGAGCGGACAAUAAAAUUUCGAGCCCAAAUGUCAUACGCAUGCAUUUGCAUAUGAUUUAGCCGGCGCUGGGUGAACAGUAUCCGUAUCCGAAUCGGAUUCGGAAU .............((((((((((..............(((.....((((((.(((......))).))))))......)))((((......)))).))))).(((.....))))))))... (-32.06 = -32.26 + 0.20)

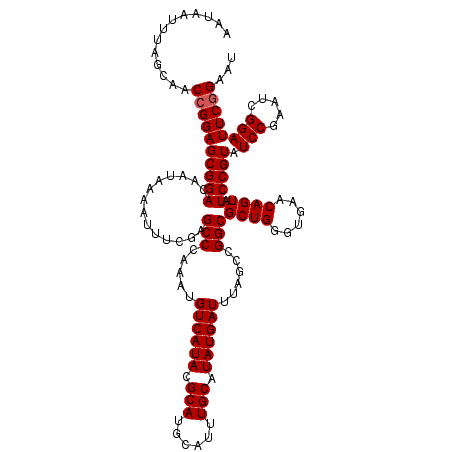
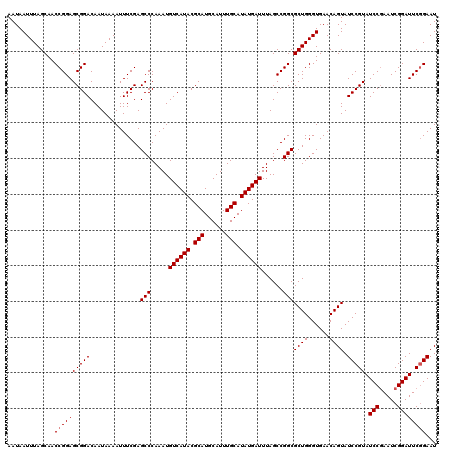
| Location | 10,168,237 – 10,168,357 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.33 |
| Mean single sequence MFE | -35.74 |
| Consensus MFE | -36.00 |
| Energy contribution | -35.68 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.50 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10168237 120 - 20766785 AUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUGGGCUCGAAAUUUUAUUGUCCGCUCCGGUUGCUAAAUUAUU ..((((.(((((...))))).))))............((((((((.....(((((((((......))))))))).....((((.............))))...)))).))))........ ( -36.42) >DroSec_CAF1 8103 120 - 1 AUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUGGGCUCGAAAUUUUAUUGUCCGCUCCGGUUGCUAAAUUAUU ..((((.(((((...))))).))))............((((((((.....(((((((((......))))))))).....((((.............))))...)))).))))........ ( -36.42) >DroSim_CAF1 7943 120 - 1 AUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUGGGCUCGAAAUUUUAUUGUCCGCUCCGGUUGCUAAAUUAUU ..((((.(((((...))))).))))............((((((((.....(((((((((......))))))))).....((((.............))))...)))).))))........ ( -36.42) >DroEre_CAF1 8101 120 - 1 AUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUGGGCUCGAAAUUUUAUUGUCCGCUCCGGUUGCUAAAUUAUU ..((((.(((((...))))).))))............((((((((.....(((((((((......))))))))).....((((.............))))...)))).))))........ ( -36.42) >DroYak_CAF1 7885 120 - 1 AUUCUGAAUCCGAUUUGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUGGGCUCGAAAUUUUAUUGUCCGCUCCGGUUGCUAAAUUAUU ..((((.((((.....)))).))))............((((((((.....(((((((((......))))))))).....((((.............))))...)))).))))........ ( -33.02) >consensus AUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUGGGCUCGAAAUUUUAUUGUCCGCUCCGGUUGCUAAAUUAUU ..((((.(((((...))))).))))............((((((((.....(((((((((......))))))))).....((((.............))))...)))).))))........ (-36.00 = -35.68 + -0.32)

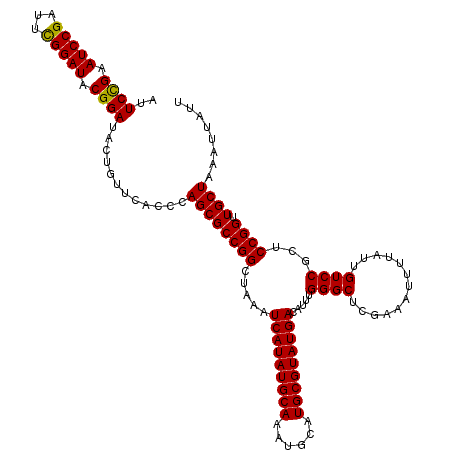

| Location | 10,168,277 – 10,168,375 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -23.08 |
| Energy contribution | -22.60 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10168277 98 - 20766785 CCUGAUUCUG----UGA------------------UUCUGAUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUG .........(----(((------------------.......((((.(((((...))))).))))......))))..(((....)))...(((((((((......)))))))))...... ( -27.12) >DroSec_CAF1 8143 98 - 1 CCUGAUUUUG----UGA------------------UUCUGAUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUG .........(----(((------------------.......((((.(((((...))))).))))......))))..(((....)))...(((((((((......)))))))))...... ( -27.12) >DroSim_CAF1 7983 98 - 1 CCUGAUUCUG----UGA------------------UUCUGAUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUG .........(----(((------------------.......((((.(((((...))))).))))......))))..(((....)))...(((((((((......)))))))))...... ( -27.12) >DroEre_CAF1 8141 102 - 1 CCCGAUCCUGCUCCUGA------------------UUCUGAUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUG .(((....((((..(((------------------.......((((.(((((...))))).))))......)))...)))).))).....(((((((((......)))))))))...... ( -27.72) >DroYak_CAF1 7925 120 - 1 CCUGACUCUGCUCCUGAUUCUGCUUCUCAUUCUGAUUCUGAUUCUGAAUCCGAUUUGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUG .(((.(.(((....(((.((((..(((...((.(((((.......))))).))...)))..))))......)))..))).).))).....(((((((((......)))))))))...... ( -26.30) >consensus CCUGAUUCUG____UGA__________________UUCUGAUUCCGAAUCCGAUUCGGAUACGGAUACUGUUCACCCAGCGCCGGCUAAAUCAUAUGCAAAUGCAUGCGUAUGACAUUUG .(((......................................((((.(((((...))))).))))..(((......)))...))).....(((((((((......)))))))))...... (-23.08 = -22.60 + -0.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:24 2006