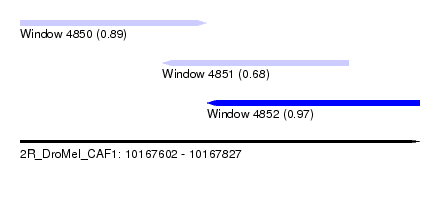
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,167,602 – 10,167,827 |
| Length | 225 |
| Max. P | 0.970256 |

| Location | 10,167,602 – 10,167,707 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.70 |
| Mean single sequence MFE | -25.76 |
| Consensus MFE | -18.56 |
| Energy contribution | -19.04 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.887117 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10167602 105 + 20766785 AUGGGAACAUACAUUUUCUUACGUUUAUUUUGAUGUGUGGCGGCAAAAGGGCGUGAAAUACGCCCAAAAAAAGGGAGACCGGAAGAGGAGGGUAAG---------------AAGCAGAGG (((....)))....((((((((.(((.(((((.((.....)).)))))(((((((...))))))).......((....)).......))).)))))---------------)))...... ( -29.40) >DroSec_CAF1 7388 87 + 1 AUGGGAAAAUACAUUUUCUUACGUUUAUUUUGAUGUGUGGCGGCAAAAGGGCGUGAAAUGCGCCCCAA-AAAGGGAGACCGGAAG--------------------------------AGG .((((((((....))))))))......(((((.((.....)).)))))((((((.....))))))...-...((....)).....--------------------------------... ( -22.00) >DroSim_CAF1 7319 104 + 1 AUGGGAAAAUACAUUUUCUUACGUUUAUUUUGAUGUGUGGCGGCAAAAGGGCGUGAAAUACGCCCCAA-AAAGGGAGACCGGAAGAGAGGGGUAAG---------------AAGCAGAGG ..............((((((((.(((.(((((.(((......)))...(((((((...))))))))))-)).((....)).......))).)))))---------------)))...... ( -26.40) >DroEre_CAF1 7493 104 + 1 ACGGGAAAAUACAUUUUCUUACGUUUAUUAUGAUGUGUGGCGGCAAAAGGGCGUGAAAUACGCCCAAA-AAAGGCAGGCCGGAAAUGGAGGGAAAG---------------AAGCAGAGG ...........(((((((.((((((......)))))).(((.((....(((((((...)))))))...-....))..)))))))))).........---------------......... ( -26.90) >DroYak_CAF1 7270 119 + 1 AUGGGGAAAUACAUUUUCUUUCGUUUAUUAUGAUGUGUGGCGGCAAAAGGGCAUGAAAUACGCCCAAA-AAAGGGAGACCGGAAAUGGAGGAAGAGCAGGGAGAGGAGGGAAAGCAGAGG .............(((((((((.(((....((..(((((.((((......)).))...)))))(((..-...((....)).....)))........))...))).)))))))))...... ( -24.10) >consensus AUGGGAAAAUACAUUUUCUUACGUUUAUUUUGAUGUGUGGCGGCAAAAGGGCGUGAAAUACGCCCAAA_AAAGGGAGACCGGAAGAGGAGGGAAAG_______________AAGCAGAGG ..(((((((....))))))).(((((((......))).))))((....((((((.....)))))).......((....)).................................))..... (-18.56 = -19.04 + 0.48)

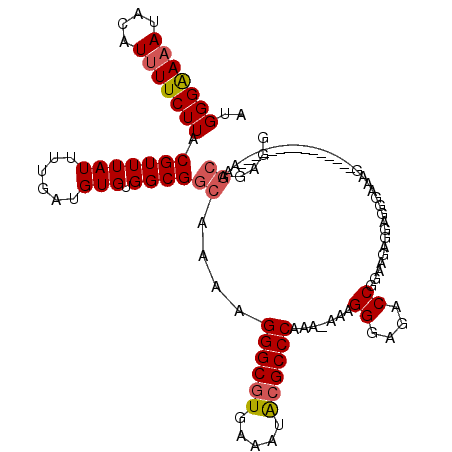
| Location | 10,167,682 – 10,167,787 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 85.87 |
| Mean single sequence MFE | -13.52 |
| Consensus MFE | -11.60 |
| Energy contribution | -11.35 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677025 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10167682 105 - 20766785 CAAUUUGACAUAAAACCUUUGCCAGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUUAUCCCUUCCCCGCCUCUGCUUCUUACCCUCCUCUUCC ................((((((((((.......))))))....((((((....))))))))))................((....)).................. ( -13.40) >DroSec_CAF1 7467 88 - 1 CAAUUUGACAUAAAACCUUUGGCGGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCACCU-----------------CUUCC .....................(((((.((.......)).)).)))((((((((((....)))))).)))).............-----------------..... ( -14.50) >DroSim_CAF1 7398 105 - 1 CAAUUUGACAUAAAACCUUUGCCGGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCGCCUCUGCUUCUUACCCCUCUCUUCC .....((((((.........((((((.......))))))....((((((....))))))...))).)))..........((....)).................. ( -14.50) >DroEre_CAF1 7572 104 - 1 CAAUUUGACAUAAAACCUUUGCUGUAUCCUUU-UUCGGCUUACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCGCCUCUGCUUCUUUCCCUCCAUUUCC ((.((((.((((((.....(((.(((.((...-...))..))))))......)))))))))).))..............((....)).................. ( -11.70) >consensus CAAUUUGACAUAAAACCUUUGCCGGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCGCCUCUGCUUCUUACCCUCCUCUUCC .....((((((.........((((((.......))))))....((((((....))))))...))).))).................................... (-11.60 = -11.35 + -0.25)

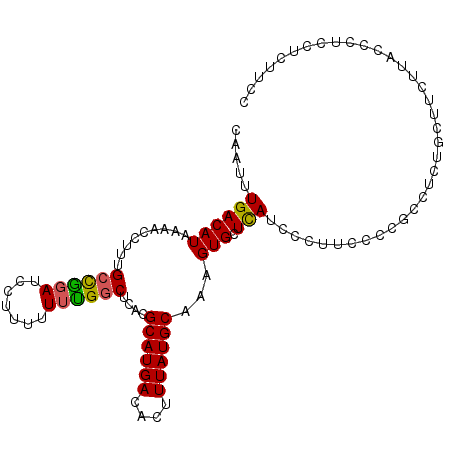
| Location | 10,167,707 – 10,167,827 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.58 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -25.86 |
| Energy contribution | -25.58 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
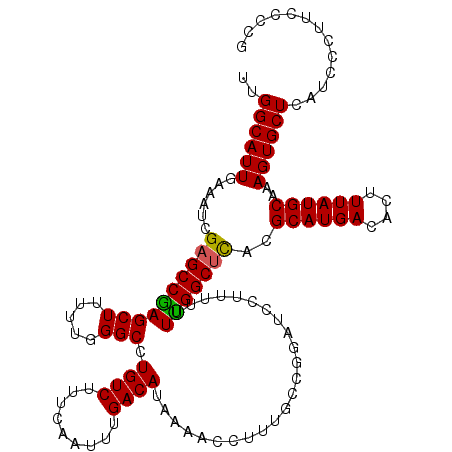
>2R_DroMel_CAF1 10167707 120 - 20766785 UUGGCAUUGAAAUCGAGCCGAGCUUUUCGGGCCUGUCUUUCAAUUUGACAUAAAACCUUUGCCAGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUUAUCCCUUCCCCG (((((.(((....))))))))......((((..((((.........))))......((((((((((.......))))))....((((((....)))))))))).............)))) ( -28.20) >DroSec_CAF1 7475 120 - 1 UUGGCAUUGAAAUCGAGCCGAGCUUUUUGGGCCUGUCUUUCAAUUUGACAUAAAACCUUUGGCGGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCA ..((((((......((((((((......((..(((((......(((......))).....)))))..))....))))))))..((((((....))))))..))))))............. ( -30.90) >DroSim_CAF1 7423 120 - 1 UUGGCAUUGAAAUCGAGCCGAGCUUUUUGGGCCUGUCUUUCAAUUUGACAUAAAACCUUUGCCGGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCG ..(((.(((....))))))((((.(((((((((((((.........)))).....((......))...........)))))..((((((....)))))))))).))))............ ( -30.50) >DroEre_CAF1 7597 119 - 1 UUGGCAUUGAAAUUGAGCCGAGCUUUUUGGGCCUGUCUUUCAAUUUGACAUAAAACCUUUGCUGUAUCCUUU-UUCGGCUUACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCG ..((((((.....(((((((((((.....))).((((.........))))......................-.)))))))).((((((....))))))..))))))............. ( -26.80) >DroYak_CAF1 7389 119 - 1 UUGGCAUUGAAAUUGAGCCAAGCUUUUUGGGCCUGUCUUUCAAUUUGACAUAAAACCUUUGUCGUAUCCUUU-UUUGGCCCACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUUCCCG ..((....((((.(((((...(((((.((((((............(((((.........)))))........-...)))))).((((((....))))))))))))))))....)))))). ( -30.30) >consensus UUGGCAUUGAAAUCGAGCCGAGCUUUUUGGGCCUGUCUUUCAAUUUGACAUAAAACCUUUGCCGGAUCCUUUUUUUGGCUCACGCAUGACACUUUAUGCAAAGUGCUCAUCCCUUCCCCG ..((((((......((((((((((.....))).((((.........))))........................)))))))..((((((....))))))..))))))............. (-25.86 = -25.58 + -0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:21 2006