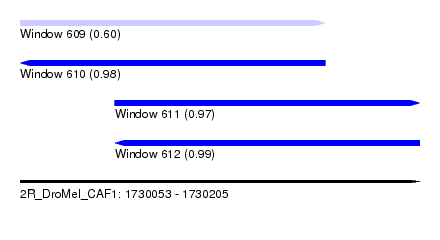
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,730,053 – 1,730,205 |
| Length | 152 |
| Max. P | 0.990091 |

| Location | 1,730,053 – 1,730,169 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -23.53 |
| Consensus MFE | -18.09 |
| Energy contribution | -18.87 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597594 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1730053 116 + 20766785 UAAUAAGAAUGUGUGCUGCAUGAAUGUAUAUGUUUCGUCUUUUUAAGAAUUAUCGUUGCUCAAUUUAUUUACAGUUGAACUGUACAGUUGAACUGUAAAUAAAUACAGUCUUUCGU ....((((.((((.((.(((((((.........)))))........((....)))).)).).((((((((((((((.((((....)))).))))))))))))))))).)))).... ( -27.10) >DroSec_CAF1 14686 103 + 1 UAAUAAGAAGUUGUGGUGCAUGAAUGUAUGUGUUUCCUCUUUUUAAGAAUUGUCGUUGCUCAAUUUAUUUACAGUUG-------------AACUGUAAAUAAAUACAGUCUUUUGG ...(((((((..(..(..((((....))))..)..)..)))))))(((.((((.........((((((((((((...-------------..)))))))))))))))))))..... ( -22.00) >DroSim_CAF1 15441 101 + 1 UAAUAAGAAGUUGUGGUGCAUGAAUGUAUGUGUUUCCUCUUUUUAAGAAUUGUAGCUGCUCAAUUUAUUUACAGUUG-------------AACUGUAAAUAAAU--AGUCUUUUGU ...(((((((..(..(..((((....))))..)..)..)))))))............(((..((((((((((((...-------------..))))))))))))--)))....... ( -21.50) >consensus UAAUAAGAAGUUGUGGUGCAUGAAUGUAUGUGUUUCCUCUUUUUAAGAAUUGUCGUUGCUCAAUUUAUUUACAGUUG_____________AACUGUAAAUAAAUACAGUCUUUUGU ...(((((((....(..((((........))))..)..)))))))............(((..((((((((((((((.((((....)))).))))))))))))))..)))....... (-18.09 = -18.87 + 0.78)

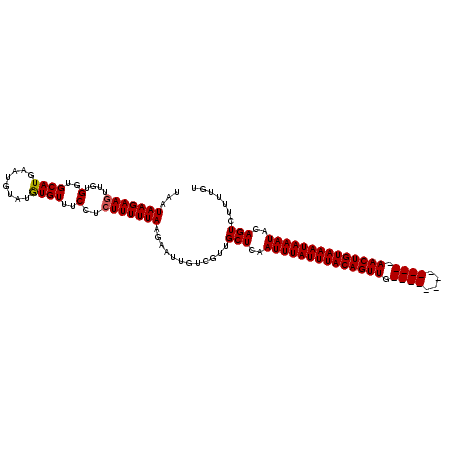
| Location | 1,730,053 – 1,730,169 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.07 |
| Mean single sequence MFE | -19.60 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1730053 116 - 20766785 ACGAAAGACUGUAUUUAUUUACAGUUCAACUGUACAGUUCAACUGUAAAUAAAUUGAGCAACGAUAAUUCUUAAAAAGACGAAACAUAUACAUUCAUGCAGCACACAUUCUUAUUA ....((((.(((((((((((((((((.((((....)))).))))))))))))))...(((..((....))..........(((.........))).))).....))).)))).... ( -23.20) >DroSec_CAF1 14686 103 - 1 CCAAAAGACUGUAUUUAUUUACAGUU-------------CAACUGUAAAUAAAUUGAGCAACGACAAUUCUUAAAAAGAGGAAACACAUACAUUCAUGCACCACAACUUCUUAUUA ....((((.(((((((((((((((..-------------...))))))))))))...(((........(((.....)))(....)...........)))...)))...)))).... ( -17.70) >DroSim_CAF1 15441 101 - 1 ACAAAAGACU--AUUUAUUUACAGUU-------------CAACUGUAAAUAAAUUGAGCAGCUACAAUUCUUAAAAAGAGGAAACACAUACAUUCAUGCACCACAACUUCUUAUUA ....(((((.--((((((((((((..-------------...)))))))))))).).(((........(((.....)))(....)...........))).........)))).... ( -17.90) >consensus ACAAAAGACUGUAUUUAUUUACAGUU_____________CAACUGUAAAUAAAUUGAGCAACGACAAUUCUUAAAAAGAGGAAACACAUACAUUCAUGCACCACAACUUCUUAUUA ....((((....((((((((((((((.((((....)))).))))))))))))))...(((........(((.....))).(((.........))).))).........)))).... (-15.30 = -15.97 + 0.67)

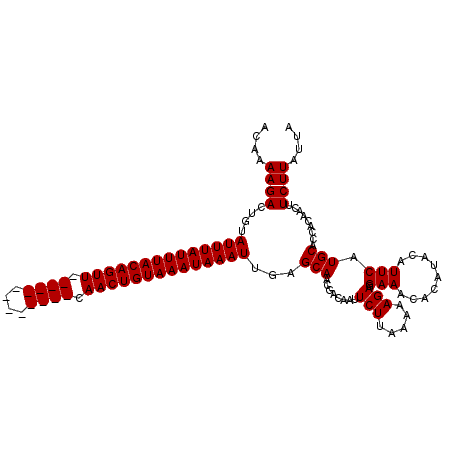
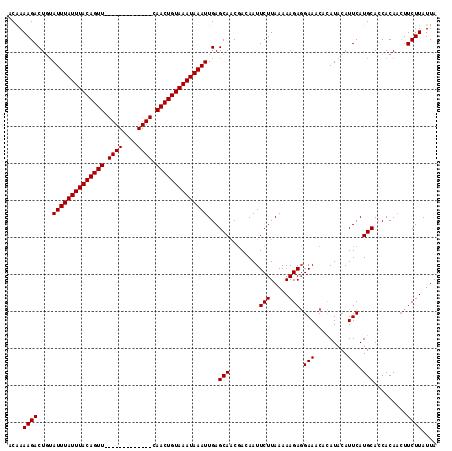
| Location | 1,730,089 – 1,730,205 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -24.67 |
| Consensus MFE | -17.90 |
| Energy contribution | -18.57 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1730089 116 + 20766785 GUCUUUUUAAGAAUUAUCGUUGCUCAAUUUAUUUACAGUUGAACUGUACAGUUGAACUGUAAAUAAAUACAGUCUUUCGUUGAA-UGGGUUUUUUUUAAUCGGUUUAUGUAGCUAAA ..................(((((...((((((((((((((.((((....)))).))))))))))))))............((((-(.((((......)))).))))).))))).... ( -29.40) >DroSec_CAF1 14722 102 + 1 CUCUUUUUAAGAAUUGUCGUUGCUCAAUUUAUUUACAGUUG-------------AACUGUAAAUAAAUACAGUCUUUUGGUGAAAUAGG--UUUUUUAUUGGGUUUGUGUAGCUAAA ..................(((((...((((((((((((...-------------..))))))))))))((((...((..((((((....--..))))))..)).))))))))).... ( -23.50) >DroSim_CAF1 15477 100 + 1 CUCUUUUUAAGAAUUGUAGCUGCUCAAUUUAUUUACAGUUG-------------AACUGUAAAUAAAU--AGUCUUUUGUUGAA-UAGU-UUUUUUUAAUGGGUUUGUGAAGCUAAA ................(((((.(.((((((((((((((...-------------..))))))))))))--.....((..(((((-....-....)))))..))..)).).))))).. ( -21.10) >consensus CUCUUUUUAAGAAUUGUCGUUGCUCAAUUUAUUUACAGUUG_____________AACUGUAAAUAAAUACAGUCUUUUGUUGAA_UAGG_UUUUUUUAAUGGGUUUGUGUAGCUAAA ..................(((((.((((((((((((((((.((((....)))).)))))))))))))).......((..(((((..........)))))..))..)).))))).... (-17.90 = -18.57 + 0.67)

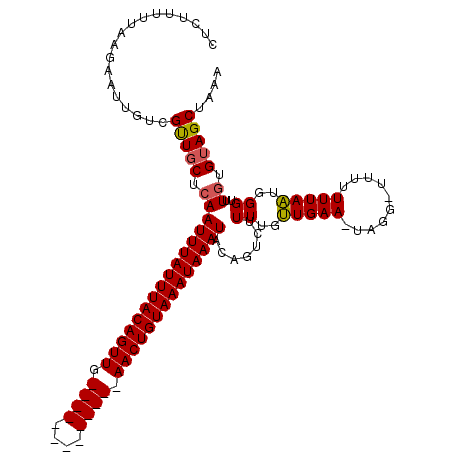
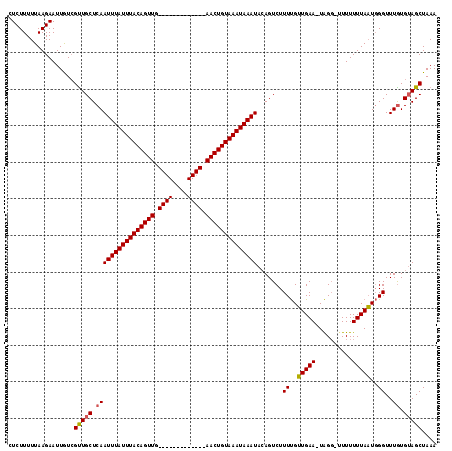
| Location | 1,730,089 – 1,730,205 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.14 |
| Mean single sequence MFE | -16.60 |
| Consensus MFE | -14.20 |
| Energy contribution | -14.87 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1730089 116 - 20766785 UUUAGCUACAUAAACCGAUUAAAAAAAACCCA-UUCAACGAAAGACUGUAUUUAUUUACAGUUCAACUGUACAGUUCAACUGUAAAUAAAUUGAGCAACGAUAAUUCUUAAAAAGAC ....(((.((......................-.....(....).....((((((((((((((.((((....)))).)))))))))))))))))))..................... ( -21.80) >DroSec_CAF1 14722 102 - 1 UUUAGCUACACAAACCCAAUAAAAAA--CCUAUUUCACCAAAAGACUGUAUUUAUUUACAGUU-------------CAACUGUAAAUAAAUUGAGCAACGACAAUUCUUAAAAAGAG ....(((.((................--......((.......))....((((((((((((..-------------...)))))))))))))))))..................... ( -12.80) >DroSim_CAF1 15477 100 - 1 UUUAGCUUCACAAACCCAUUAAAAAAA-ACUA-UUCAACAAAAGACU--AUUUAUUUACAGUU-------------CAACUGUAAAUAAAUUGAGCAGCUACAAUUCUUAAAAAGAG ..(((((.(.(((..............-....-.((.......))..--.(((((((((((..-------------...)))))))))))))).).)))))................ ( -15.20) >consensus UUUAGCUACACAAACCCAUUAAAAAAA_CCUA_UUCAACAAAAGACUGUAUUUAUUUACAGUU_____________CAACUGUAAAUAAAUUGAGCAACGACAAUUCUUAAAAAGAG ....(((.((............................(....).....((((((((((((((.((((....)))).)))))))))))))))))))..................... (-14.20 = -14.87 + 0.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:47 2006