| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,161,886 – 10,162,003 |
| Length | 117 |
| Max. P | 0.708293 |

| Location | 10,161,886 – 10,162,003 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -29.45 |
| Consensus MFE | -19.58 |
| Energy contribution | -21.34 |
| Covariance contribution | 1.76 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598270 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161886 117 + 20766785 AAGUAAGUAAUGGCAAUGGCAAUGGCUAUGGAAAUGACAGCAUGCCAAUGGAAAUCUGCUUACAAUUUUUAUGCCAGGCCAUUCCAAAUUUGAUUGCCAGCCAACC---AAAGGCCCGGU ..........(((.((((((..((((.((((((((...((((..((...)).....))))....)))))))))))).))))))))).........(((.(((....---...)))..))) ( -31.80) >DroSec_CAF1 1689 113 + 1 AAGUAAGUAAUGGCAAUGGCAAUGUC------AAUGGCAGCAUGCCAAUGGAAAUCUGCUUACAAUUUUUAUGCCAGACCAUUCCAAAUUUGAUUGCCAGCCAACC-CCAUUGGCUCGGU ..........(((((((((((.(((.------....)))...))))..(((((.(((((.............).))))...)))))......)))))))(((((..-...)))))..... ( -29.92) >DroSim_CAF1 1616 113 + 1 AAGUAAGUAAUGGCAAUGGCAAUGUC------AAUGGCAGCAUGCCAAUGGAAAUCUGCUUACAAUUUUUAUGCCAGACCAUUCCAAAUUUGAUUGCCAGCCAACC-CCAUUGGCUCGGU ..........(((((((((((.(((.------....)))...))))..(((((.(((((.............).))))...)))))......)))))))(((((..-...)))))..... ( -29.92) >DroEre_CAF1 1684 98 + 1 UAGUAAGUAAUGGCACUG------------------GCAGCAUGCCAGUGGAAAUCUGCUUACAAUU-UUAUGCCAGACCGUUCCAAAUUUGAUUGCCAGCCA---ACCAUUGGCUCGGU ..(((((((((..(((((------------------((.....)))))))...)).)))))))....-...........................((((((((---.....))))).))) ( -29.00) >DroYak_CAF1 1654 102 + 1 UAGUAAGUAAUGGCAAUG------------------GCAGCAUGCCAAUGGAAAUCUGCUUACAAUUUUUAUGCCAAACCAUUCCAAAUUUGAUUGCCAGCCAGCCACCAUUGGCUCGGU ..........((((((((------------------((.....)))..(((((....((.............)).......)))))......)))))))((((((((....))))).))) ( -26.62) >consensus AAGUAAGUAAUGGCAAUGGCAAUG_C______AAUGGCAGCAUGCCAAUGGAAAUCUGCUUACAAUUUUUAUGCCAGACCAUUCCAAAUUUGAUUGCCAGCCAACC_CCAUUGGCUCGGU ..........(((((((.................((((.....)))).(((((.(((((.............).))))...)))))......)))))))(((((......)))))..... (-19.58 = -21.34 + 1.76)

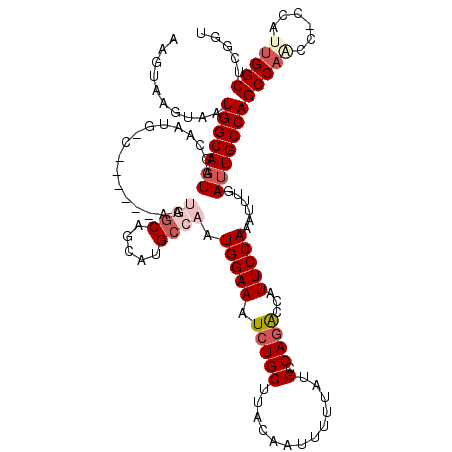
| Location | 10,161,886 – 10,162,003 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.94 |
| Mean single sequence MFE | -31.56 |
| Consensus MFE | -22.46 |
| Energy contribution | -23.38 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161886 117 - 20766785 ACCGGGCCUUU---GGUUGGCUGGCAAUCAAAUUUGGAAUGGCCUGGCAUAAAAAUUGUAAGCAGAUUUCCAUUGGCAUGCUGUCAUUUCCAUAGCCAUUGCCAUUGCCAUUACUUACUU .((((((((((---(((((.....))))))))........)))))))..........(((((..(((...((.(((((.(((((.......)))))...))))).))..))).))))).. ( -33.50) >DroSec_CAF1 1689 113 - 1 ACCGAGCCAAUGG-GGUUGGCUGGCAAUCAAAUUUGGAAUGGUCUGGCAUAAAAAUUGUAAGCAGAUUUCCAUUGGCAUGCUGCCAUU------GACAUUGCCAUUGCCAUUACUUACUU ..........(((-(..((((((((((((((...(((((..((((((((.......)))...)))))))))).((((.....))))))------))..))))))..))))...))))... ( -32.20) >DroSim_CAF1 1616 113 - 1 ACCGAGCCAAUGG-GGUUGGCUGGCAAUCAAAUUUGGAAUGGUCUGGCAUAAAAAUUGUAAGCAGAUUUCCAUUGGCAUGCUGCCAUU------GACAUUGCCAUUGCCAUUACUUACUU ..........(((-(..((((((((((((((...(((((..((((((((.......)))...)))))))))).((((.....))))))------))..))))))..))))...))))... ( -32.20) >DroEre_CAF1 1684 98 - 1 ACCGAGCCAAUGGU---UGGCUGGCAAUCAAAUUUGGAACGGUCUGGCAUAA-AAUUGUAAGCAGAUUUCCACUGGCAUGCUGC------------------CAGUGCCAUUACUUACUA ....(((((.....---)))))(((..........((((..((((((((...-...)))...)))))))))((((((.....))------------------)))))))........... ( -30.60) >DroYak_CAF1 1654 102 - 1 ACCGAGCCAAUGGUGGCUGGCUGGCAAUCAAAUUUGGAAUGGUUUGGCAUAAAAAUUGUAAGCAGAUUUCCAUUGGCAUGCUGC------------------CAUUGCCAUUACUUACUA .((.(((((....))))))).((((((.......(((((..(((((.((.......)))))))....))))).((((.....))------------------)))))))).......... ( -29.30) >consensus ACCGAGCCAAUGG_GGUUGGCUGGCAAUCAAAUUUGGAAUGGUCUGGCAUAAAAAUUGUAAGCAGAUUUCCAUUGGCAUGCUGCCAUU______G_CAUUGCCAUUGCCAUUACUUACUU ....(((((........)))))((((((......(((((..((((((((.......)))...)))))))))).((((.....)))).................))))))........... (-22.46 = -23.38 + 0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:10 2006