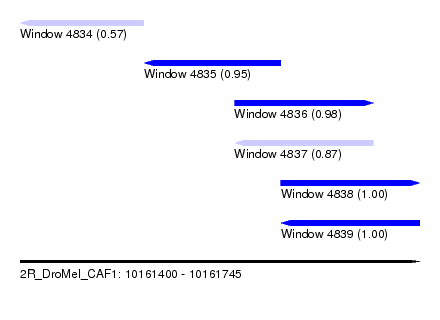
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,161,400 – 10,161,745 |
| Length | 345 |
| Max. P | 0.999369 |

| Location | 10,161,400 – 10,161,507 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 86.43 |
| Mean single sequence MFE | -26.22 |
| Consensus MFE | -18.57 |
| Energy contribution | -19.13 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.566036 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161400 107 - 20766785 AGCGUAAAGGGUAAAAUGCUGAAUUGU---UAAAUAACUAAUGCAUGGCCAAUGAACUUGCCGAGCAAAAGUUUUAGCAAUUUAUCUGCGUGCCCCCUUUUGCGCCUGCU .((((((((((....((((((((((((---((((..(((..(((.((((..........)))).)))..)))))))))))))))...))))....))))))))))..... ( -36.70) >DroSec_CAF1 1200 109 - 1 AGCGUAAAGGGUAAAAUGCUGCAUUGUAACUAAAUAACUAAUGCAUGGCCAAUGAACUUGCCGAGCAAAAGUUUUACCAAUUUAUCUGCGUGCCCCCUUUAUC-CCUGCU (((((((((((....(((((((((((............)))))))(((.....((((((((...)))..)))))..)))........))))...)))))))).-...))) ( -23.80) >DroSim_CAF1 1130 106 - 1 AGCGUAAAGGGUAAAAUGCUGAAUUGU---UAAAUAACUAAUGCAUGGCCAAUGAACUUGCCGAGCAAAAGUUUUAGCAAUUUAUCUGCGNNNNNNNNNNNNN-NNNNNN ..(((...(((((((.(((((((....---......(((..(((.((((..........)))).)))..)))))))))).)))))))))).............-...... ( -20.50) >DroEre_CAF1 1198 105 - 1 AGCGUAAAGGGUAAAAUGCUGAAUUGU---UAAAUAACUAAUGCACGGCCAAUGAACUUGCAGAGCAAAAGUUUUAGCAAUUUAUCUGCGUGCCCCUUUUUCC-C-UGCU ((((.((((((....((((((((((((---((((..(((..(((.(.((..........)).).)))..)))))))))))))))...))))..))))))....-.-)))) ( -25.70) >DroYak_CAF1 1169 105 - 1 AGCGUAAAGGGUAAAAUGCUGAAUUGU---UAAAUAACUAAUGCAUGGGCAAUGAACUUGC-GAGCGAAAGUUUUAGCAAUUUAUCUGCGCACCCCUUUUUCC-CCGGCC ..((.((((((....((((......((---(....)))....))))(.((((((((.((((-((((....))))..))))))))).))).)..))))))....-.))... ( -24.40) >consensus AGCGUAAAGGGUAAAAUGCUGAAUUGU___UAAAUAACUAAUGCAUGGCCAAUGAACUUGCCGAGCAAAAGUUUUAGCAAUUUAUCUGCGUGCCCCCUUUUCC_CCUGCU .((((...(((((((.((((((.............((((..(((.(((.(((.....)))))).)))..)))))))))).)))))))))))................... (-18.57 = -19.13 + 0.56)

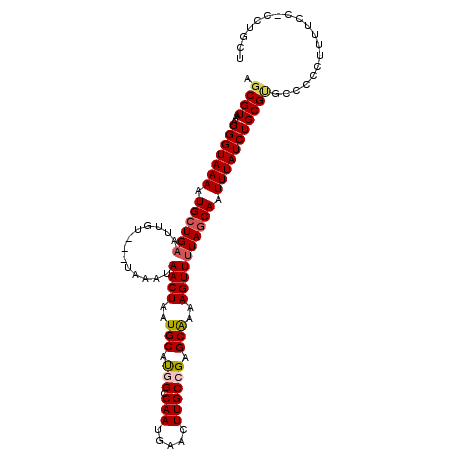
| Location | 10,161,507 – 10,161,625 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -20.44 |
| Energy contribution | -20.28 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947221 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161507 118 - 20766785 AUCUAACCAGCCACUCGCCGGGUAAAAGCAAAAGUUUAAAUUACUUUCUAGUAAAACUUUGAUCCGCAACAAAAAAAAAAUGGGCUAAAAGCAGAAUGCCAGCGAGAUUAAUGCUUAA ......((.((.....)).))....((((((((((((....((((....)))))))))))(((((((..((.........))(((............))).))).))))..))))).. ( -22.30) >DroSec_CAF1 1309 117 - 1 AUCUAACCAUCCACUCGCCGGGUAAAAGCAAAAGUUUAAAUUACUUUCUAGUGAAACUUUGAUCCGCAACAAAAA-AAAAUGGGCUAAAAGCAGAAUGCCAGCGAGAUUAAUGCUUAA ........((((.......))))..(((((((((((....(((((....)))))))))))(((((((..((....-....))(((............))).))).))))..))))).. ( -22.10) >DroSim_CAF1 1236 117 - 1 AUCUAACCAUCCACUCGCCGGGUAAAAGCAAAAGUUUAAAUUACUUUCUAGUGAAACUUUGAUCCGCAACAAAAA-AAAAUGGGCUAAAAGCUGAAUGCCAGCGAGAUUAAUGCUUAA ........((((.......))))..(((((((((((....(((((....)))))))))))(((((((..((....-....))(((............))).))).))))..))))).. ( -22.10) >DroEre_CAF1 1303 115 - 1 AUCUAACCAGGCACUCGCAGGGUAAAAGCAAAAGUUUAAAUUACUUUCUAGUGAAACUUUGAUCCGCAACAAA---AAAAUAGGCUAAAAGCAGAUUGCCAGCGAGAUUAAUGCUUAA ........((((((((((..(((((..((.((((((....(((((....))))))))))).....))......---.......((.....))...))))).))))).....))))).. ( -23.70) >DroYak_CAF1 1274 116 - 1 AUCUAUCCAGCCACUCGCAGGGUAAAAGCAAAAGUUUAAAUUACUUUCUAGUGAAACUUUGAUCCGCAACAAAA--AAAAUAGGCUAAAAGCAGAUUGCCAGCGAGAUUAAUGCUUAA ...(((((.((.....)).))))).(((((((((((....(((((....)))))))))))(((((((.......--..(((..((.....))..)))....))).))))..))))).. ( -22.32) >consensus AUCUAACCAGCCACUCGCCGGGUAAAAGCAAAAGUUUAAAUUACUUUCUAGUGAAACUUUGAUCCGCAACAAAAA_AAAAUGGGCUAAAAGCAGAAUGCCAGCGAGAUUAAUGCUUAA .............(((((.((((.......((((((....(((((....))))))))))).)))).................(((............))).)))))............ (-20.44 = -20.28 + -0.16)


| Location | 10,161,585 – 10,161,705 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -24.70 |
| Energy contribution | -25.30 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.79 |
| SVM RNA-class probability | 0.977485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161585 120 + 20766785 UUUAAACUUUUGCUUUUACCCGGCGAGUGGCUGGUUAGAUUUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUU .((((((.(((((.((((.(((((.....))))).))))................((((......))))((((((..((......))..))))))....)))))....))))))...... ( -28.80) >DroSec_CAF1 1386 120 + 1 UUUAAACUUUUGCUUUUACCCGGCGAGUGGAUGGUUAGAUUUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUCAACACUUU ....(((.(((((.......((((((((((((((..((((......)))).))))))).))))..))).((((((..((......))..))))))....)))))....)))......... ( -28.90) >DroSim_CAF1 1313 120 + 1 UUUAAACUUUUGCUUUUACCCGGCGAGUGGAUGGUUAGAUUUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUU .((((((.(((((.......((((((((((((((..((((......)))).))))))).))))..))).((((((..((......))..))))))....)))))....))))))...... ( -32.30) >DroEre_CAF1 1378 120 + 1 UUUAAACUUUUGCUUUUACCCUGCGAGUGCCUGGUUAGAUUUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUU .((((((.(((((..........(((((...(((..((((......)))).)))....)))))......((((((..((......))..))))))....)))))....))))))...... ( -26.60) >DroYak_CAF1 1350 120 + 1 UUUAAACUUUUGCUUUUACCCUGCGAGUGGCUGGAUAGAUUUUUACAUUUUGCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUU .((((((.(((((............(((((((((((.((..........))..))))))))...)))..((((((..((......))..))))))....)))))....))))))...... ( -25.90) >consensus UUUAAACUUUUGCUUUUACCCGGCGAGUGGCUGGUUAGAUUUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUU .((((((.(((((..........(((((((.(((..((((......)))).))).))).))))......((((((..((......))..))))))....)))))....))))))...... (-24.70 = -25.30 + 0.60)

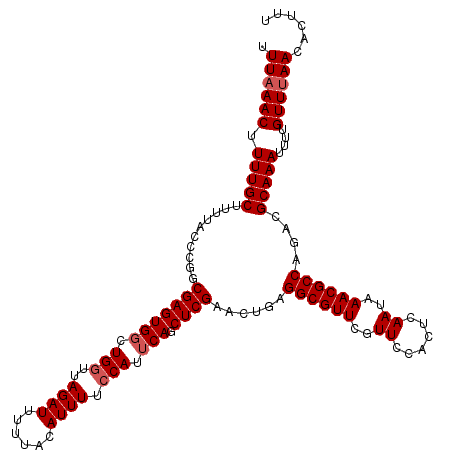
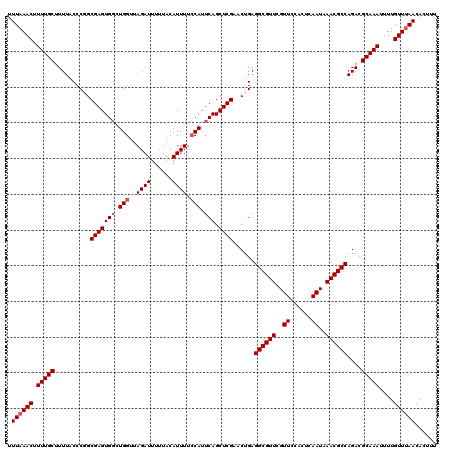
| Location | 10,161,585 – 10,161,705 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -30.08 |
| Consensus MFE | -24.81 |
| Energy contribution | -25.85 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161585 120 - 20766785 AAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAAAUCUAACCAGCCACUCGCCGGGUAAAAGCAAAAGUUUAAA ......((((((....(((((.((((((.......(((((...(((((......))))).((((..((((...........))))..))))))))))))))).....))))).)))))). ( -31.61) >DroSec_CAF1 1386 120 - 1 AAAGUGUUGAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAAAUCUAACCAUCCACUCGCCGGGUAAAAGCAAAAGUUUAAA ......((((((....(((((.((((((.......(((((((.(((((......))))).......(((.................)))))))))))))))).....))))).)))))). ( -30.24) >DroSim_CAF1 1313 120 - 1 AAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAAAUCUAACCAUCCACUCGCCGGGUAAAAGCAAAAGUUUAAA ......((((((....(((((.((((((.......(((((((.(((((......))))).......(((.................)))))))))))))))).....))))).)))))). ( -30.14) >DroEre_CAF1 1378 120 - 1 AAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAAAUCUAACCAGGCACUCGCAGGGUAAAAGCAAAAGUUUAAA ......((((((....(((((.(((((((((..((......))..))))))(((((....)))))..)))...............(((..((....))..)))....))))).)))))). ( -28.50) >DroYak_CAF1 1350 120 - 1 AAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGCAAAAUGUAAAAAUCUAUCCAGCCACUCGCAGGGUAAAAGCAAAAGUUUAAA ......((((((....(((((((..((((((..((......))..))))))(((((....))))))))))))...........(((((.((.....)).))))).........)))))). ( -29.90) >consensus AAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAAAUCUAACCAGCCACUCGCCGGGUAAAAGCAAAAGUUUAAA ......((((((....(((((.((((((.......((((((..(((((......)))))..(((..((((...........))))..))))))))))))))).....))))).)))))). (-24.81 = -25.85 + 1.04)

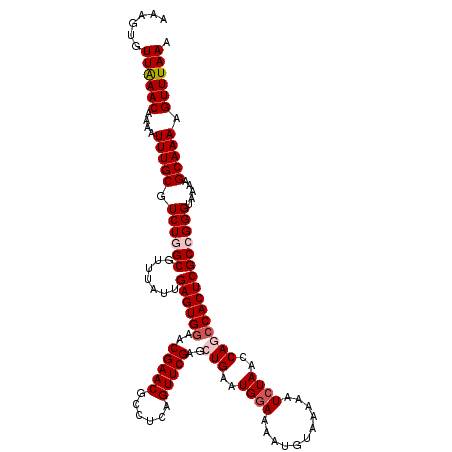
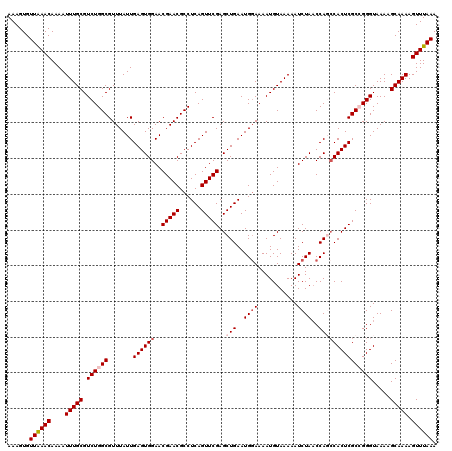
| Location | 10,161,625 – 10,161,745 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -27.25 |
| Energy contribution | -27.09 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995623 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161625 120 + 20766785 UUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUUUUGCCUAUUGGCAUUGUAAAAUGGGAAUAAAAUGCGAAGA ...............((((......))))((((((..((......))..))))))...((((..((((((((..((.(((.((((....))))....))).)).)))))))))))).... ( -27.20) >DroSec_CAF1 1426 120 + 1 UUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUCAACACUUUUUGCCUAUUGGCAUUGUAAAAUGGGAAUAAAAUGCGAAGA ...............((((......))))((((((..((......))..))))))...((((..((((((((..((.(((.((((....))))....))).)).)))))))))))).... ( -29.10) >DroSim_CAF1 1353 120 + 1 UUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUUUUGCCUAUUGGCAUUGUAAAAUGGGAAUAAAAUGCGAAGA ...............((((......))))((((((..((......))..))))))...((((..((((((((..((.(((.((((....))))....))).)).)))))))))))).... ( -27.20) >DroEre_CAF1 1418 120 + 1 UUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUUUUGCCUAUUGGCAUUGUAAAAUAGGAAUAAAAUGCGAAGG ...........((..((((......))))((((((..((......))..))))))...((((..((((((((.........((((....))))...........))))))))))))..)) ( -27.25) >DroYak_CAF1 1390 120 + 1 UUUUACAUUUUGCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUUUUGCCUAUUGGCAUUGUAAAAUAAGAAUAAAAUGCGAAGA (((..((((((((..((((......))))((((((..((......))..))))))....))...((((((((.(((.....((((....)))).))).))))))))..))))))..))). ( -30.20) >consensus UUUUACAUUUUCCAUUCAGCUCGAACUGAGGCGUUCGUUCCACUCAAUAAACGCCAGACGCAAAUUUUGUUUAACACUUUUUGCCUAUUGGCAUUGUAAAAUGGGAAUAAAAUGCGAAGA ...............((((......))))((((((..((......))..))))))...((((..((((((((.........((((....))))...........)))))))))))).... (-27.25 = -27.09 + -0.16)


| Location | 10,161,625 – 10,161,745 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -31.04 |
| Energy contribution | -30.64 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10161625 120 - 20766785 UCUUCGCAUUUUAUUCCCAUUUUACAAUGCCAAUAGGCAAAAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAA .....(((((((((((((((((...(((((((..(.(((((...((.....))...))))).).)))))))....))))))..(((((......)))))....)))))..)))))).... ( -32.00) >DroSec_CAF1 1426 120 - 1 UCUUCGCAUUUUAUUCCCAUUUUACAAUGCCAAUAGGCAAAAAGUGUUGAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAA .....(((((((((((((((((...(((((((..(.(((((...((.....))...))))).).)))))))....))))))..(((((......)))))....)))))..)))))).... ( -32.00) >DroSim_CAF1 1353 120 - 1 UCUUCGCAUUUUAUUCCCAUUUUACAAUGCCAAUAGGCAAAAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAA .....(((((((((((((((((...(((((((..(.(((((...((.....))...))))).).)))))))....))))))..(((((......)))))....)))))..)))))).... ( -32.00) >DroEre_CAF1 1418 120 - 1 CCUUCGCAUUUUAUUCCUAUUUUACAAUGCCAAUAGGCAAAAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAA (((((((......((((.((((...(((((((..(.(((((...((.....))...))))).).)))))))....))))))))(((((......))))).)).))).))........... ( -29.90) >DroYak_CAF1 1390 120 - 1 UCUUCGCAUUUUAUUCUUAUUUUACAAUGCCAAUAGGCAAAAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGCAAAAUGUAAAA ...................(((((((.((((....)))).................(((((((..((((((..((......))..))))))(((((....)))))))))))).))))))) ( -29.80) >consensus UCUUCGCAUUUUAUUCCCAUUUUACAAUGCCAAUAGGCAAAAAGUGUUAAACAAAAUUUGCGUCUGGCGUUUAUUGAGUGGAACGAACGCCUCAGUUCGAGCUGAAUGGAAAAUGUAAAA .....(((((((((((((((((...(((((((..(.(((((...((.....))...))))).).)))))))....))))))..(((((......)))))....)))))..)))))).... (-31.04 = -30.64 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:08 2006