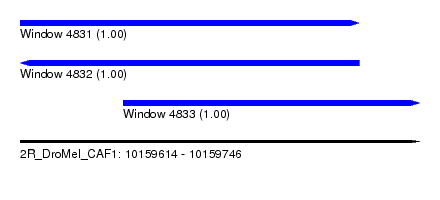
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,159,614 – 10,159,746 |
| Length | 132 |
| Max. P | 0.999993 |

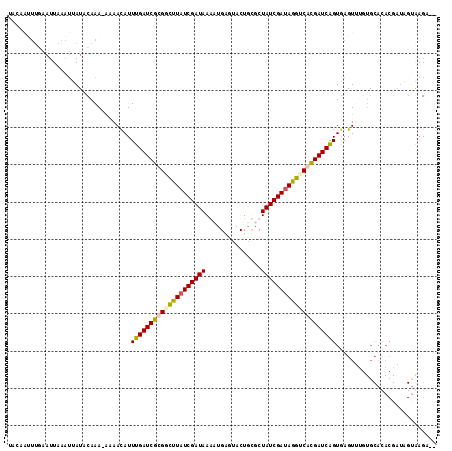
| Location | 10,159,614 – 10,159,726 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 71.45 |
| Mean single sequence MFE | -30.88 |
| Consensus MFE | -22.71 |
| Energy contribution | -22.96 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.80 |
| Structure conservation index | 0.74 |
| SVM decision value | 5.78 |
| SVM RNA-class probability | 0.999993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10159614 112 + 20766785 UACUACUUGAAUUAAAUUAUUCAAA-AAAACAUUUGAUCGCGGCUUAUCGAUAAGCUGAUUACUGUGCUAUCGAUAGGUCAUGAUCAGUAAGUAGCUGCACACGAUAGCAAGA-- ..((((((((((......))))...-.......(((((((.((((((((((((.((..........)))))))))))))).))))))).)))))).(((........)))...-- ( -33.70) >DroSec_CAF1 21789 113 + 1 UGCAAUUUGAAUUAAAUUAUACAAAAAAAACAUUUGAUCGUGUCUUAUCGAUAAAAUGAGUACUGCGCUAUCGAUAGGUCACGAUCAGUGAGUUUGUGCACACGAUAGUGAGA-- ((((.((((.((......)).))))..((((..(((((((((.((((((((((...............)))))))))).)))))))))...)))).))))(((....)))...-- ( -32.36) >DroSim_CAF1 15063 102 + 1 ----------UAUAUAAUAUACAAA-ACAAUAUUUGAUCGUGUCUUAUCGAUAAAAUGAGUACUGCGCUAUCGAUAGGUCACGAUCAGUGAGUUUGUGCACACGAUAGUGAGA-- ----------.........((((((-.......(((((((((.((((((((((...............)))))))))).)))))))))....))))))..(((....)))...-- ( -30.46) >DroYak_CAF1 16804 99 + 1 AAGAGUUUAAAUUAAAUUAUUCAAAAGCAAUAUUUGAUCGCGGUAUAUCGAUAAGCUGAUAACUGUGCUAUCGACAUGCCACGAUCGGUGAGU----------------AAGUAU ................(((((((..........(((((((.(((((.((((((.((..........)))))))).))))).))))))))))))----------------)).... ( -27.00) >consensus UACAAUUUGAAUUAAAUUAUACAAA_AAAACAUUUGAUCGCGGCUUAUCGAUAAAAUGAGUACUGCGCUAUCGAUAGGUCACGAUCAGUGAGUUUGUGCACACGAUAGUAAGA__ .................................((((((((((((((((((((...............))))))))))))))))))))........................... (-22.71 = -22.96 + 0.25)


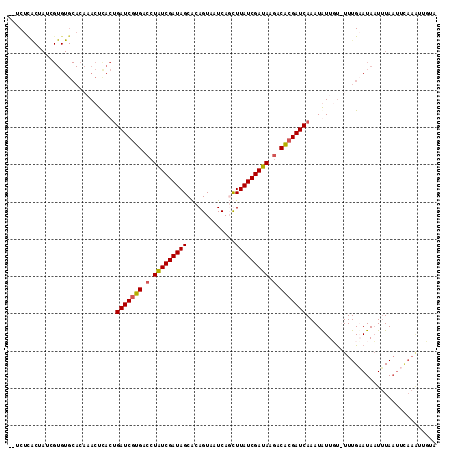
| Location | 10,159,614 – 10,159,726 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.45 |
| Mean single sequence MFE | -26.31 |
| Consensus MFE | -16.88 |
| Energy contribution | -16.95 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.64 |
| SVM decision value | 5.31 |
| SVM RNA-class probability | 0.999983 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10159614 112 - 20766785 --UCUUGCUAUCGUGUGCAGCUACUUACUGAUCAUGACCUAUCGAUAGCACAGUAAUCAGCUUAUCGAUAAGCCGCGAUCAAAUGUUUU-UUUGAAUAAUUUAAUUCAAGUAGUA --...(((........)))(((((((..(((((.((.(.((((((((((..........)).)))))))).).)).)))))........-...((((......))))))))))). ( -27.00) >DroSec_CAF1 21789 113 - 1 --UCUCACUAUCGUGUGCACAAACUCACUGAUCGUGACCUAUCGAUAGCGCAGUACUCAUUUUAUCGAUAAGACACGAUCAAAUGUUUUUUUUGUAUAAUUUAAUUCAAAUUGCA --...........((((((.((((....((((((((.(.(((((((((.............))))))))).).))))))))...))))....))))))................. ( -23.92) >DroSim_CAF1 15063 102 - 1 --UCUCACUAUCGUGUGCACAAACUCACUGAUCGUGACCUAUCGAUAGCGCAGUACUCAUUUUAUCGAUAAGACACGAUCAAAUAUUGU-UUUGUAUAUUAUAUA---------- --......(((.(((((((.((((....((((((((.(.(((((((((.............))))))))).).))))))))......))-))))))))).)))..---------- ( -26.72) >DroYak_CAF1 16804 99 - 1 AUACUU----------------ACUCACCGAUCGUGGCAUGUCGAUAGCACAGUUAUCAGCUUAUCGAUAUACCGCGAUCAAAUAUUGCUUUUGAAUAAUUUAAUUUAAACUCUU ......----------------.......((((((((.(((((((((((..........)).))))))))).))))))))..........(((((((......)))))))..... ( -27.60) >consensus __UCUCACUAUCGUGUGCACAAACUCACUGAUCGUGACCUAUCGAUAGCACAGUAAUCAGCUUAUCGAUAAGACACGAUCAAAUAUUGU_UUUGAAUAAUUUAAUUCAAAUUGUA .............................(((((((.(.(((((((((.............))))))))).).)))))))................................... (-16.88 = -16.95 + 0.06)


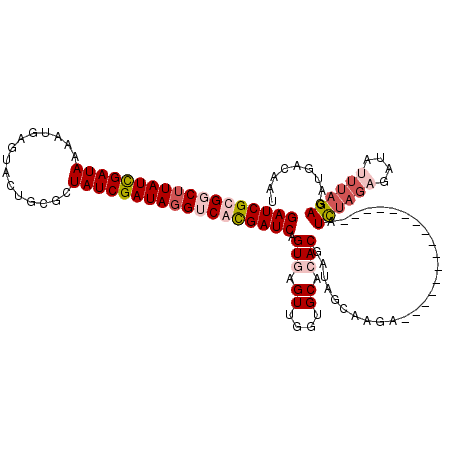
| Location | 10,159,648 – 10,159,746 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -21.92 |
| Energy contribution | -24.36 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.71 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10159648 98 + 20766785 GAUCGCGGCUUAUCGAUAAGCUGAUUACUGUGCUAUCGAUAGGUCAUGAUCAGUAAGUAGCUGCACACGAUAGCAAGA----------------AUUC------AUCUAGAGAUGACAAU (((((.((((((((((((.((..........)))))))))))))).)))))........(((((....).))))....----------------..((------(((....))))).... ( -31.10) >DroSec_CAF1 21824 104 + 1 GAUCGUGUCUUAUCGAUAAAAUGAGUACUGCGCUAUCGAUAGGUCACGAUCAGUGAGUUUGUGCACACGAUAGUGAGA----------------GUCUAGAGAUAUUUAGAGAUGACAAU (((((((.((((((((((...............)))))))))).))))))).(((.((....)).)))....((.(..----------------.((((((....))))))..).))... ( -31.86) >DroSim_CAF1 15087 104 + 1 GAUCGUGUCUUAUCGAUAAAAUGAGUACUGCGCUAUCGAUAGGUCACGAUCAGUGAGUUUGUGCACACGAUAGUGAGA----------------GUCUAGAGAUAUUUAGAGAUGACAAU (((((((.((((((((((...............)))))))))).))))))).(((.((....)).)))....((.(..----------------.((((((....))))))..).))... ( -31.86) >DroEre_CAF1 21408 120 + 1 GAUCGCGGCUUAUUGAUAACUUGAUUACUUUACUAUCGAUAUGUCACGAUCAGUGAGUGGUCGCAAACGAUAACAAGUACUUAUCGCAUCACAGAUCUAGCGAUAUUUAGAGAUGACAAU .((((((((.((((((((...(((.....))).)))))))).)))..((((.(((((((((.....((........))....))))).)))).))))..)))))................ ( -28.30) >consensus GAUCGCGGCUUAUCGAUAAAAUGAGUACUGCGCUAUCGAUAGGUCACGAUCAGUGAGUUGGUGCACACGAUAGCAAGA________________AUCUAGAGAUAUUUAGAGAUGACAAU ((((((((((((((((((...............)))))))))))))))))).(((.((....)).)))...........................((((((....))))))......... (-21.92 = -24.36 + 2.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:02 2006