| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,152,664 – 10,152,774 |
| Length | 110 |
| Max. P | 0.969969 |

| Location | 10,152,664 – 10,152,774 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 72.53 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -20.57 |
| Energy contribution | -20.94 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.65 |
| SVM RNA-class probability | 0.969969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10152664 110 + 20766785 UUCGCCCUUCGUUUGGCUGCGAGCAUUAACCUCAGAAUGCAUUUGCAGCUGAAAAACUUCAAAAGGGAG-CAAGGAGGAGAGCGUCGAAAGGGGG-G-GG--GGGGGGGGAAG-UG (((.((((((.((..((((((((((((........)))))..)))))))..))...((((....(....-)...........(.((....)).))-)-))--)))))).))).-.. ( -36.70) >DroPse_CAF1 8344 105 + 1 ----C------AGUGGCUGCGAGCAAUAACCUCAGAAUGCAUUUACAGCUGCUGUGCUGUGGACAGGAG-GACAGAGGAGCAGGAUGCGGUUGACUGGUAUGGGCCGGAGAGCCAG ----.------..(((((........(((((.((...(((.((((((((......))))))))..(...-..)......)))...)).))))).(((((....)))))..))))). ( -33.20) >DroSec_CAF1 13133 111 + 1 UUCGCCCUUCGUUUGGCUGCGAGCAUUAACCUCAGAAUGCAUUUGCAGCUCGGAAAGUCCA-GAGGGGG-CAAGGAGGAGAAGGAGGAGAGGGAC-G-AAAAGUGCGGAAAAG-UG ((((((((((.((((((((((((((((........)))))..))))))).))))..((((.-....)))-)........))))).........((-.-....)))))))....-.. ( -33.00) >DroSim_CAF1 8062 111 + 1 UUCGCCCUUCGUUUGGCUGCGAGCAUUAACCUCAGAAUGCAUUUGCAGCUCGGAAACUCCA-GAGGGGG-CAAGGAGGAGAAGGAGGAGAGGGGC-G-AAAAGUGCGGAAAAG-UG ((((((((((.((((((((((((((((........)))))..))))))).))))..((((.-.......-...))))...........)))))))-)-)).............-.. ( -39.00) >DroEre_CAF1 13399 107 + 1 UUCGCCCUUCGUUUGGCUGCGAGCAUUAACCUCAGAAUGCAUUUGCAGCUGGAAAACUCCG-GAGGGGGACAGGGGG---AAGGAGGUAAGGGG----AAUGGAGGGGAAAAG-UG (((.(((((((((((((((((((((((........)))))..))))))))......((((.-...(....).)))).---.............)----))))))))))))...-.. ( -38.40) >DroYak_CAF1 8136 99 + 1 UUCGCCCUUCGUUUGGCUGCGAGCAUUAACCUCAGAAUGCAUUUGCAGCUGGAAAACUCCG-GAGGGGUUCAAGGAG---AA--------UGGG----AAUGGAGGGGAAAAG-UG (((.(((((((((((((((((((((((........)))))..))))))))......((((.-...........))))---..--------...)----))))))))))))...-.. ( -35.90) >consensus UUCGCCCUUCGUUUGGCUGCGAGCAUUAACCUCAGAAUGCAUUUGCAGCUGGAAAACUCCA_GAGGGGG_CAAGGAGGAGAAGGAGGAGAGGGGC_G_AAUGGAGCGGAAAAG_UG ....(((((..((..((((((((((((........)))))..)))))))..))...((((.............))))...........)))))....................... (-20.57 = -20.94 + 0.37)

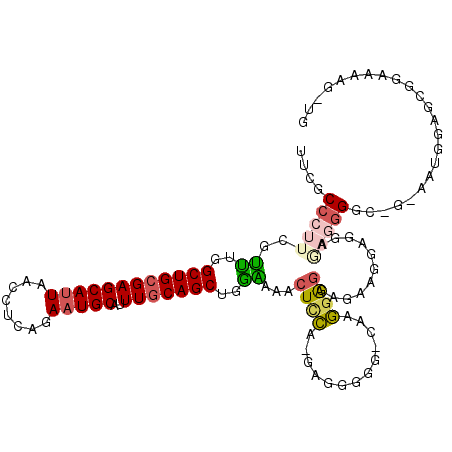
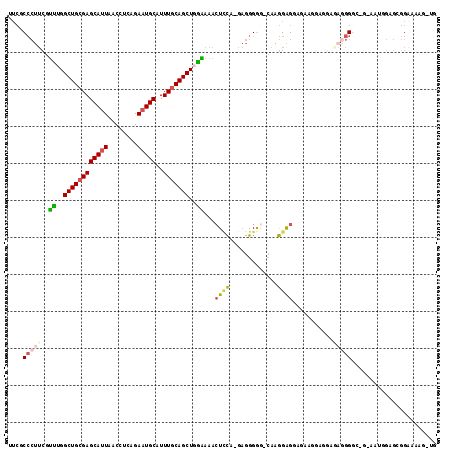
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:43:00 2006