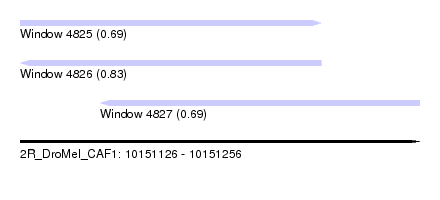
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,151,126 – 10,151,256 |
| Length | 130 |
| Max. P | 0.827730 |

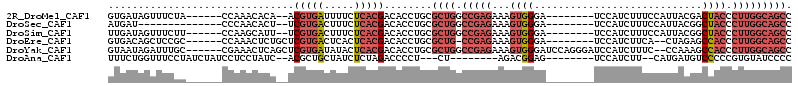
| Location | 10,151,126 – 10,151,224 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 65.85 |
| Mean single sequence MFE | -25.89 |
| Consensus MFE | -11.36 |
| Energy contribution | -13.31 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.44 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.685093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10151126 98 + 20766785 GUGAUAGUUUCUA------CCAAACACA--ACGUGAUUUUCUCACGACACCUGCGCUGGCCGAGAAAGUGGGA--------UCCAUCUUUCCAUUACGACUACCCUUGGCAGCC (((...((((...------..))))...--.(((((.....))))).)))....((((.(((((..(((((((--------.......)))))))..(.....)))))))))). ( -24.00) >DroSec_CAF1 11586 90 + 1 AUGAU--------------CCCAACACU--UCGUGACUUUCUCACGACACCUGCGCUGGCCGAGAAAGUGGGA--------UCCAUCUUUCCAUUACGGCUACCCUUGGCAGCC ..(((--------------(((..(((.--..)))((((((((......((......))..))))))))))))--------))..............((((.((...)).)))) ( -25.40) >DroSim_CAF1 6567 98 + 1 UUGAUAGUUUCUU------CCAAGCAUU--UCGUGACUUUCUCACGACACCUGCGCUGGCCGAGAAAGUGGGA--------UCCAUCUUUCCAUUACGGCUACCCUUGGCAGCC .............------....(((..--((((((.....))))))....)))((((.(((((..(((((((--------.......)))))))..((...))))))))))). ( -26.90) >DroEre_CAF1 11882 97 + 1 GUGACAGCUCCGC------CCAAACUCUGCUCGUGACUCACUCACGACACCUGCGCUG-CCGAGAAAGUGGGA--------UCCAUCUUCA--CUAGAGCCACCCUUGGCAGCC (((...((.....------.........))((((((.....)))))))))....((((-(((((...((((..--------....(((...--..))).)))).))))))))). ( -28.74) >DroYak_CAF1 6635 106 + 1 GUAAUAGAUUUGC------CGAAACUCAGCUCGUGAUAUACUCACGACACCUGCGCUGGCCGAGAAAGUGGGAUCCAGGGAUCCAUCUUUC--CCAAAGCCACCCUUGGCAGCC .........((((------(((..(.(((.((((((.....))))))...))).).((((.(.(((((..(((((....)))))..)))))--.)...))))...))))))).. ( -33.80) >DroAna_CAF1 7556 91 + 1 UUUCUGGUUUCCUAUCUAUCCUCCUAUC--ACGCUGCUAUCUCUAGACCCCU---CU--------AGACGGAG--------UCCAUCUU--CAUGAUGUCCCCCGUGUAUCCCC ............................--((((.......((((((....)---))--------))).((.(--------..((((..--...))))..).))))))...... ( -16.50) >consensus GUGAUAGUUUCGC______CCAAACACU__UCGUGACUUUCUCACGACACCUGCGCUGGCCGAGAAAGUGGGA________UCCAUCUUUCCAUUACGGCCACCCUUGGCAGCC ...............................(((((.....)))))........((((.(((((...((((............................)))).))))))))). (-11.36 = -13.31 + 1.94)

| Location | 10,151,126 – 10,151,224 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 65.85 |
| Mean single sequence MFE | -32.50 |
| Consensus MFE | -14.81 |
| Energy contribution | -16.25 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10151126 98 - 20766785 GGCUGCCAAGGGUAGUCGUAAUGGAAAGAUGGA--------UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAAUCACGU--UGUGUUUGG------UAGAAACUAUCAC (((((((...)))))))......(((((.(((.--------..))))))))((.(((((((((((((((....))....))).)--)))).))))------).))......... ( -30.50) >DroSec_CAF1 11586 90 - 1 GGCUGCCAAGGGUAGCCGUAAUGGAAAGAUGGA--------UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAGUCACGA--AGUGUUGGG--------------AUCAU (((((((...))))))).....((((((.(((.--------..)))))))))...(((((((.....((((((.....))))))--.))))))).--------------..... ( -34.30) >DroSim_CAF1 6567 98 - 1 GGCUGCCAAGGGUAGCCGUAAUGGAAAGAUGGA--------UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAGUCACGA--AAUGCUUGG------AAGAAACUAUCAA (((((((...)))))))......(((((.(((.--------..))))))))((..((((.(((....((((((.....))))))--..)))))))------..))......... ( -34.20) >DroEre_CAF1 11882 97 - 1 GGCUGCCAAGGGUGGCUCUAG--UGAAGAUGGA--------UCCCACUUUCUCGG-CAGCGCAGGUGUCGUGAGUGAGUCACGAGCAGAGUUUGG------GCGGAGCUGUCAC (((..((...))..)))....--....(((((.--------((((((((...(((-((.(...).))))).))))).(((.(((((...))))))------))))).))))).. ( -32.10) >DroYak_CAF1 6635 106 - 1 GGCUGCCAAGGGUGGCUUUGG--GAAAGAUGGAUCCCUGGAUCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGUAUAUCACGAGCUGAGUUUCG------GCAAAUCUAUUAC ((((((((....))))...((--(((((..(((((....)))))..))))))))))).....((((.((((((.....))))))((((.....))------))..))))..... ( -41.40) >DroAna_CAF1 7556 91 - 1 GGGGAUACACGGGGGACAUCAUG--AAGAUGGA--------CUCCGUCU--------AG---AGGGGUCUAGAGAUAGCAGCGU--GAUAGGAGGAUAGAUAGGAAACCAGAAA .......(((((((..((((...--..))))..--------)))).(((--------((---(....)))))).........))--)...............(....)...... ( -22.50) >consensus GGCUGCCAAGGGUAGCCGUAAUGGAAAGAUGGA________UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAGUCACGA__AGUGUUUGG______AAGAAACUAUCAC ((((((.....))))))..........(((((............(((((.(.(.....).).)))))((((((.....)))))).......................))))).. (-14.81 = -16.25 + 1.44)
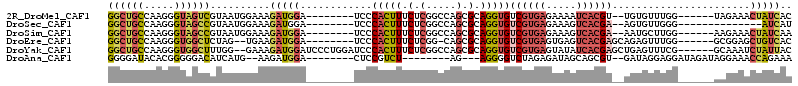

| Location | 10,151,152 – 10,151,256 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 77.63 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -21.65 |
| Energy contribution | -21.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.688195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10151152 104 - 20766785 AGAGUCGACUAUUUGAUGGCUUUUGGGGGGCUGGCUGCCAAGGGUAGUCGUAAUGGAAAGAUGGA--------UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAAUC ...........(((.(((((.((((.(.(((((((((((...))))))).....((((((.(((.--------..))))))))).))))..).)))).))))).)))..... ( -32.60) >DroSec_CAF1 11604 103 - 1 AGAGUCGACUAUUUGAUGGCUUUUGGG-GGCCGGCUGCCAAGGGUAGCCGUAAUGGAAAGAUGGA--------UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAGUC ......((((.(((.(((((.((((.(-(((((((((((...))))))).....((((((.(((.--------..))))))))).))))..).)))).))))).))).)))) ( -41.00) >DroSim_CAF1 6593 103 - 1 AGAGUCGACUAUUUGAUGGCUUUUGGG-GGCUGGCUGCCAAGGGUAGCCGUAAUGGAAAGAUGGA--------UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAGUC ......((((.(((.(((((.((((.(-(((((((((((...))))))).....((((((.(((.--------..))))))))).))))..).)))).))))).))).)))) ( -38.00) >DroEre_CAF1 11910 100 - 1 AGAGUCGACUAUUUGAUGGCUUUUGGG-GGCCGGCUGCCAAGGGUGGCUCUAG--UGAAGAUGGA--------UCCCACUUUCUCGG-CAGCGCAGGUGUCGUGAGUGAGUC .....((((.(((((.((((((....)-)))))((((((.(((((((.((((.--......))))--------..)))))..)).))-)))).))))))))).......... ( -35.00) >DroYak_CAF1 6663 109 - 1 AGAGUCGACUAUUUGAUGGCUUUUGUG-GGCUGGCUGCCAAGGGUGGCUUUGG--GAAAGAUGGAUCCCUGGAUCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGUAUAUC (((((((.(.....).)))))))..((-.(((((((((((....))))...((--(((((..(((((....)))))..)))))))))))))).))((((((....).))))) ( -45.40) >DroAna_CAF1 7588 89 - 1 CGAGUCGACUUCUUGAUGGCCUU-GGG-GUCUGGGGAUACACGGGGGACAUCAUG--AAGAUGGA--------CUCCGUCU--------AG---AGGGGUCUAGAGAUAGCA .(((((.(((((.(((((.((((-(..-(((....)))...)))))..))))).)--))).).))--------)))..(((--------((---(....))))))....... ( -32.70) >consensus AGAGUCGACUAUUUGAUGGCUUUUGGG_GGCUGGCUGCCAAGGGUAGCCGUAAUGGAAAGAUGGA________UCCCACUUUCUCGGCCAGCGCAGGUGUCGUGAGAAAGUC .....((((.(((((.(((((.......(((((.((.....)).)))))......(((((.(((...........))))))))..)))))...))))))))).......... (-21.65 = -21.77 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:57 2006