| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,149,265 – 10,149,369 |
| Length | 104 |
| Max. P | 0.795483 |

| Location | 10,149,265 – 10,149,369 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.94 |
| Mean single sequence MFE | -23.63 |
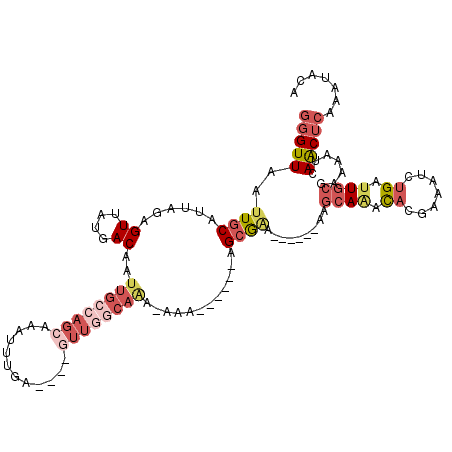
| Consensus MFE | -8.45 |
| Energy contribution | -10.70 |
| Covariance contribution | 2.25 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.56 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
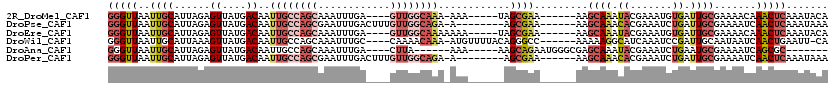
>2R_DroMel_CAF1 10149265 104 - 20766785 GGGUUAAUUGCAUUAGAGUUAUGACAAUUGCCAGCAAAUUUGA----GUUGGCAAA-AAA-----UAGCGAA------AAGCAAAUACGAAAUGUGAUUGCGAAAACAAACUCAAAUACA (((((............(((((.....((((((((........----)))))))).-..)-----))))...------..((((.(((.....))).)))).......)))))....... ( -24.20) >DroPse_CAF1 5392 105 - 1 GGGUUAAUUGCAUUAGAGUUAUGACAAUUGCCAGCGAAUUUGACUUUGUUGGCAGA-A--------AGCGAA------AAGCAAACACGAAAUCUGAUUGCGAAAAUCAACUCAAAUAAA (((((..((((((((((.(..((....(((((((((((......))))))))))).-.--------.((...------..))...))..)..))))).))))).....)))))....... ( -27.40) >DroEre_CAF1 10035 105 - 1 GGGUUAAUUGCAUUAGAGUUAUGACAAUUGCCAGCAAAUUUGA----GUUGGCAAAAAAA-----UAGCGAA------AAGCAAAUACGAAAUGUGAUUGCGAAAACAAACUCAAAUACA (((((............(((((.....((((((((........----))))))))....)-----))))...------..((((.(((.....))).)))).......)))))....... ( -24.60) >DroWil_CAF1 17399 108 - 1 GGGUUAAUUGCAUUAAAGUUAUGACAAUUGCCAGCAAAUUUGC----CAAAACAAA-AUGUUUUACAGGGCC------AAAAAGGCAUCAAAUCCGAUUGCAAUAAUCAACUGAAUU-CA .(((.((((((((.......))).)))))))).(((....)))----.(((((...-..))))).(((.(((------.....))).........(((((...)))))..)))....-.. ( -18.70) >DroAna_CAF1 4737 98 - 1 GGGUUAAUUGCAUUAGAGUUAUGACAAUUGCCAGCAAAUUUGA----CUUA------AAA-----AAGCAGAAUGGGCGAGCAAAUACGAAAUCUGAAUGCGAAAAUCAGCGC------- .((((..((((((((((((....))..((((..((..((((..----(((.------...-----)))..))))..))..))))........)))).)))))).)))).....------- ( -19.50) >DroPer_CAF1 5422 105 - 1 GGGUUAAUUGCAUUAGAGUUAUGACAAUUGCCAGCGAAUUUGACUUUGUUGGCAGA-A--------AGCGAA------AAGCAAACACGAAAUCUGAUUGCGAAAAUCAACUCAAAUAAA (((((..((((((((((.(..((....(((((((((((......))))))))))).-.--------.((...------..))...))..)..))))).))))).....)))))....... ( -27.40) >consensus GGGUUAAUUGCAUUAGAGUUAUGACAAUUGCCAGCAAAUUUGA____GUUGGCAAA_AAA______AGCGAA______AAGCAAACACGAAAUCUGAUUGCGAAAAUCAACUCAAAUACA (((((..((((......((....))..((((((((............))))))))............)))).........((((.((.......)).)))).......)))))....... ( -8.45 = -10.70 + 2.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:54 2006