
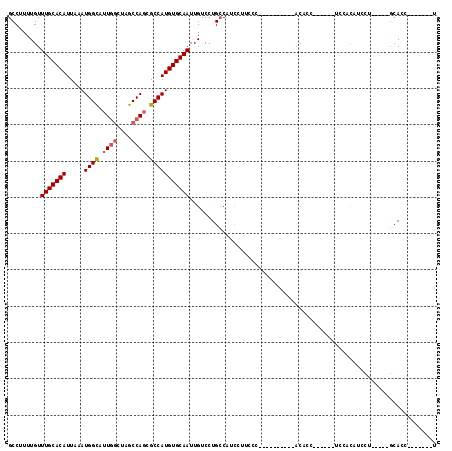
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,146,716 – 10,146,841 |
| Length | 125 |
| Max. P | 0.982919 |

| Location | 10,146,716 – 10,146,816 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 77.50 |
| Mean single sequence MFE | -19.79 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10146716 100 - 20766785 GCCUUUUGUUUGCACAUUAAAUGGCAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCAUCCUUCCCACACCUUCCCACACC------UUCACAUCCU-----GCACC-------U ((.......(((((((.....((((.((((....)))).))))))))))).......)).........................------..........-----.....-------. ( -19.64) >DroSec_CAF1 7005 90 - 1 GCCUUUUGUUUGCACAUUAAAUGGUAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCCUCCAUCCC----------ACACC------UCCACAUCCU-----GCACC-------U ((.......(((((((.....(((..((((....))))..)))))))))).......))..........----------.....------..........-----.....-------. ( -18.64) >DroSim_CAF1 2188 90 - 1 GCCUUUUGUUUGCACAUUAAAUGGCAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCCUCCAUCCC----------ACACC------UCCACAUCCU-----GCACC-------U ((.......(((((((.....((((.((((....)))).))))))))))).......))..........----------.....------..........-----.....-------. ( -19.64) >DroYak_CAF1 2239 96 - 1 GCCUUUUGUUUGCACAUUAAAUGGCAUUCGCUAGCCACCGCCAUGUGCAAUUGUCCUGGCACUUUUCCC----------ACUCCUCAUACUCCUCAUACU-----CAACC-------U (((......((((((((....((((........)))).....)))))))).......))).........----------.....................-----.....-------. ( -20.12) >DroAna_CAF1 2117 103 - 1 GCCUUUUGUUUGCACAUUAAAUGGCAUUG-CCCGCCAGCGCCAUGUGCAAUUGUCCUGCCGGCUUUGCU----------ACCUCUU----UCUUUAUCCUUUUUGGCACCCACACCAG (((......((((((((....((((....-...)))).....))))))))..(((.....)))......----------.......----..............)))........... ( -20.90) >consensus GCCUUUUGUUUGCACAUUAAAUGGCAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCAUCCUUCCC__________ACACC______UCCACAUCCU_____GCACC_______U .........(((((((.....((((.((((....)))).))))))))))).................................................................... (-15.52 = -15.96 + 0.44)


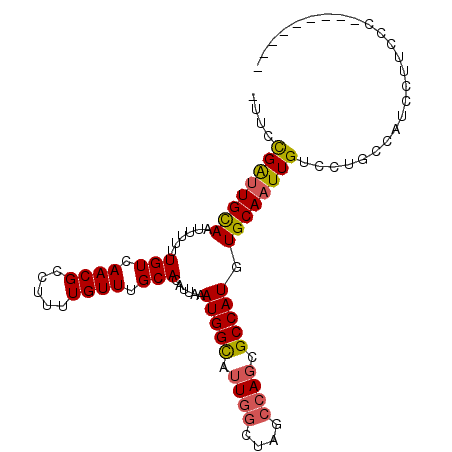
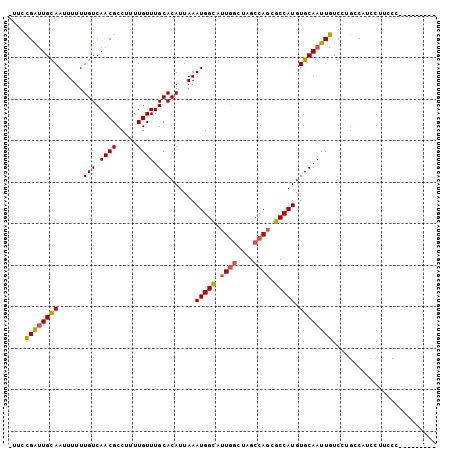
| Location | 10,146,738 – 10,146,841 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 86.02 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -19.95 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.795087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10146738 103 - 20766785 GUAACGAUUGCAAUUUUUUGUCAACGCCUUUUGUUUGCACAUUAAAUGGCAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCAUCCUUCCCACACCUUCC ((((((((((((......(((.((((.....)))).)))......(((((.((((....)))).))))).)))))))))..)))................... ( -25.20) >DroSec_CAF1 7026 94 - 1 UUUCCGAUUGCAAUUUUUUGUCAACGCCUUUUGUUUGCACAUUAAAUGGUAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCCUCCAUCCC--------- ....((((((((......(((.((((.....)))).)))......((((..((((....))))..)))).))))))))................--------- ( -22.40) >DroSim_CAF1 2209 94 - 1 UUUCCGAUUGCAAUUUUUUGUCAACGCCUUUUGUUUGCACAUUAAAUGGCAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCCUCCAUCCC--------- ....((((((((......(((.((((.....)))).)))......(((((.((((....)))).))))).))))))))................--------- ( -23.40) >DroEre_CAF1 7582 91 - 1 ---CCGAUUGCAAUUUUUUGUCAACGCCUUUUGUUUGCACAUUAAAUGGCAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGGCACCCUGCCC--------- ---.((((((((......(((.((((.....)))).)))......(((((.((((....)))).))))).))))))))....(((.....))).--------- ( -27.10) >DroYak_CAF1 2266 91 - 1 ---CCGAUUGCAAUUUUUUGUCAACGCCUUUUGUUUGCACAUUAAAUGGCAUUCGCUAGCCACCGCCAUGUGCAAUUGUCCUGGCACUUUUCCC--------- ---..(.((((((....))).))))(((......((((((((....((((........)))).....)))))))).......))).........--------- ( -21.92) >DroAna_CAF1 2152 93 - 1 GAUAUGGCUGUAAUUUUUUGUCAACGCCUUUUGUUUGCACAUUAAAUGGCAUUG-CCCGCCAGCGCCAUGUGCAAUUGUCCUGCCGGCUUUGCU--------- .(((((((.((.......(((.((((.....)))).))).......((((....-...)))))))))))))((((..(((.....))).)))).--------- ( -22.00) >consensus _UUCCGAUUGCAAUUUUUUGUCAACGCCUUUUGUUUGCACAUUAAAUGGCAUUGGCUAGCCAGCGCCAUGUGCAAUUGUCCUGCCAUCCUUCCC_________ ....((((((((......(((.((((.....)))).)))......(((((.((((....)))).))))).))))))))......................... (-19.95 = -20.07 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:53 2006