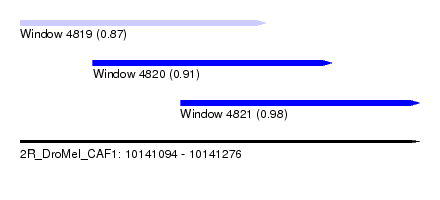
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,141,094 – 10,141,276 |
| Length | 182 |
| Max. P | 0.978584 |

| Location | 10,141,094 – 10,141,206 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -30.98 |
| Energy contribution | -30.55 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10141094 112 + 20766785 GGUUGCUCACGUAAUA------UAACUCUCUGA-AAGGGGAAUCCCUUUGAAGAUUCCACUCGCAGGACUCGAGCGCUCACUUUCCCCAUCCCGUGUCCUGCGAAGAGUAUUCAAAGGC .(((((....))))).------....(((((..-..)))))...((((((((.((((...(((((((((.((....................)).))))))))).)))).)))))))). ( -37.75) >DroSim_CAF1 6214 112 + 1 GGUUACUCACGUAAUA------UAACUCUCUGA-AAGGGGAAUCCCUUUGAAGAUUCCACUCGCAGGACUCGAGCGCUCACUUUCCCCAUCCCGUGUCCUGUGAAGAGUAUUCAAAGGC .(((((....))))).------....(((((..-..)))))...((((((((.((((...(((((((((.((....................)).))))))))).)))).)))))))). ( -35.85) >DroYak_CAF1 6239 119 + 1 GGUUAUUCACGUAAUACGAGUACAACUCUCUAAAAAGGGGAAUCCCUUUGAACAUUCCACUCGCAGGACUUGAGCGCUCACUUUCCCCAUUCUGUGUCCUGCUAAGAGCAUUCAAAGGC ...(((((.........)))))....(((((.....)))))...((((((((..(((.....(((((((..(((...............)))...)))))))...)))..)))))))). ( -25.86) >consensus GGUUACUCACGUAAUA______UAACUCUCUGA_AAGGGGAAUCCCUUUGAAGAUUCCACUCGCAGGACUCGAGCGCUCACUUUCCCCAUCCCGUGUCCUGCGAAGAGUAUUCAAAGGC .(((((....)))))...........(((((.....)))))...((((((((.((((...(((((((((.((....................)).))))))))).)))).)))))))). (-30.98 = -30.55 + -0.43)


| Location | 10,141,127 – 10,141,236 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -30.47 |
| Consensus MFE | -27.34 |
| Energy contribution | -27.35 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908031 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10141127 109 + 20766785 AAUCCCUUUGAAGAUUCCACUCGCAGGACUCGAGCGCUCACUUUCCCCAUCCCGUGUCCUGCGAAGAGUAUUCAAAGGCAAACUUUGACCGCUUUUUGCCAGGACCUAC ..(((.((((((.((((...(((((((((.((....................)).))))))))).)))).))))))((((((............)))))).)))..... ( -32.55) >DroSim_CAF1 6247 109 + 1 AAUCCCUUUGAAGAUUCCACUCGCAGGACUCGAGCGCUCACUUUCCCCAUCCCGUGUCCUGUGAAGAGUAUUCAAAGGCAAACUUUGACCGCUUUUUGCGAGGACCUAC ....((((((((.((((...(((((((((.((....................)).))))))))).)))).))))))))...........(((.....)))......... ( -31.55) >DroYak_CAF1 6279 109 + 1 AAUCCCUUUGAACAUUCCACUCGCAGGACUUGAGCGCUCACUUUCCCCAUUCUGUGUCCUGCUAAGAGCAUUCAAAGGCAAACUUUGACCGCUUUUUGCCAGGACCUAC ......................((((((...(((......)))......))))))(((((((.((((((..((((((.....))))))..)))))).).)))))).... ( -27.30) >consensus AAUCCCUUUGAAGAUUCCACUCGCAGGACUCGAGCGCUCACUUUCCCCAUCCCGUGUCCUGCGAAGAGUAUUCAAAGGCAAACUUUGACCGCUUUUUGCCAGGACCUAC ....((((((((.((((...(((((((((.((....................)).))))))))).)))).))))))))..........((((.....))..))...... (-27.34 = -27.35 + 0.01)

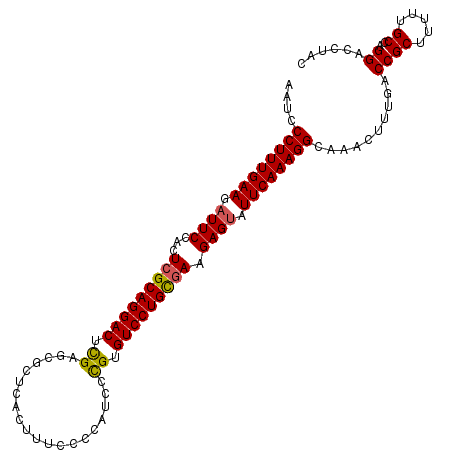
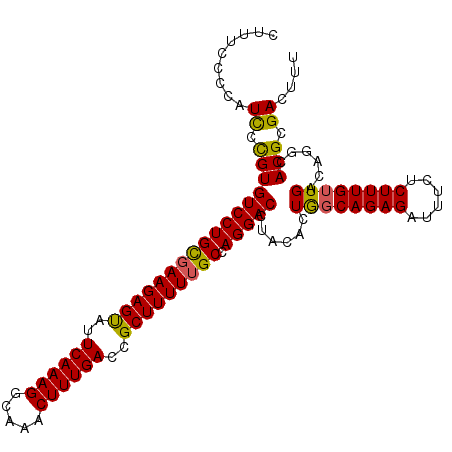
| Location | 10,141,167 – 10,141,276 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -31.10 |
| Energy contribution | -30.67 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978584 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10141167 109 + 20766785 CUUUCCCCAUCCCGUGUCCUGCGAAGAGUAUUCAAAGGCAAACUUUGACCGCUUUUUGCCAGGACCUACACUGGCAGAGAUUUCUCUUUGUUGACAGGCACGCGACUUU ............((((((((((((((((...((((((.....))))))....((((((((((........))))))))))...)))))))....)))).)))))..... ( -37.90) >DroSim_CAF1 6287 109 + 1 CUUUCCCCAUCCCGUGUCCUGUGAAGAGUAUUCAAAGGCAAACUUUGACCGCUUUUUGCGAGGACCUACACUAGCAGAGAUUUCUCUUUGUUGACAGGCACGCGACUUU .........((.(((((((((..((((((..((((((.....))))))..))))))..).))))(((....(((((((((....)))))))))..))))))).)).... ( -34.60) >DroYak_CAF1 6319 109 + 1 CUUUCCCCAUUCUGUGUCCUGCUAAGAGCAUUCAAAGGCAAACUUUGACCGCUUUUUGCCAGGACCUACACUGCCAGAGAUUUCGCUUUGUUGACAAUCACGCGACUUU .........(((((.(((((((.((((((..((((((.....))))))..)))))).).)))))).........)))))...((((.(((....)))....)))).... ( -28.70) >consensus CUUUCCCCAUCCCGUGUCCUGCGAAGAGUAUUCAAAGGCAAACUUUGACCGCUUUUUGCCAGGACCUACACUGGCAGAGAUUUCUCUUUGUUGACAGGCACGCGACUUU .........((.(((((((((((((((((..((((((.....))))))..))))))))).)))))......((((((((......))))))))......))).)).... (-31.10 = -30.67 + -0.43)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:51 2006