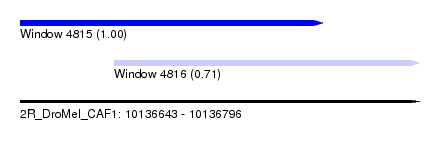
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,136,643 – 10,136,796 |
| Length | 153 |
| Max. P | 0.999647 |

| Location | 10,136,643 – 10,136,759 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.64 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -34.00 |
| Energy contribution | -33.45 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.01 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999647 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
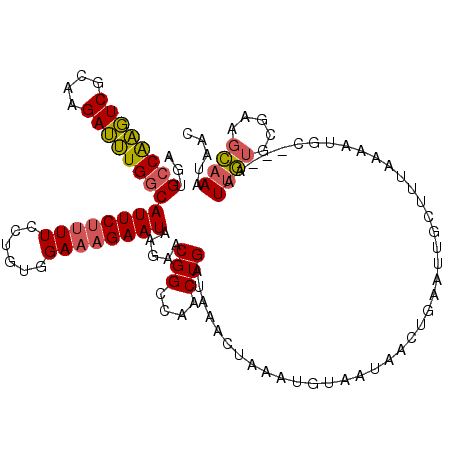
>2R_DroMel_CAF1 10136643 116 + 20766785 ---UAGUUACUUGAUGAUUGUUC-UUUUUCUGAAGAGUCGAGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUAGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUUUAACAACU ---(((((....(.((..(((((-((((((((..(((..((((((((((((....))))))))))))..)))...))))))....)))))))..)).).....)))))............ ( -37.40) >DroSec_CAF1 1961 116 + 1 ---UAGUUACUUGAUGAUUGUUC-UUUUUCUGAAGAGUCGAGUGCCAAGUCGCAAGAUUUGUCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAAUUAAAUGUAAUAACU ---..(((((....((..(((((-((.((((........(((((.((((((....)))))).)))))..((((...)))))))).)))))))..))..(((....)))...))))).... ( -28.30) >DroSim_CAF1 1962 116 + 1 ---UAGUUACUUGAUGAUUGUUC-UUUUUCUGAAGAGUCGAGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUGUAAUAACU ---(((((....(.((..(((((-((.((((........((((((((((((....))))))))))))..((((...)))))))).)))))))..)).).....)))))............ ( -37.30) >DroEre_CAF1 1980 117 + 1 ---UAGUUUCUUGAUGAUUGUUCUAUUUUCUGAAGAGUCGAGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAACCUAAAUGUAAUGACA ---....((((((..((((.(((........))).))))((((((((((((....))))))))))))((((((...))))))..))))))(((....))).................... ( -34.40) >DroYak_CAF1 1970 116 + 1 ---UAGUUUCUUGAUGAUUGUUC-AUAUUCUGAAGAGUCGAGUGCCAAGUCGCAAGAUUUGGCAUUCUAUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUCUAAUAACA ---((((((...(.((..(((((-.((((((........((((((((((((....))))))))))))..((((...)))))))))).)))))..)).)....))))))............ ( -38.30) >DroAna_CAF1 2030 115 + 1 GUUUGAUUCUUUGAUGAAUGUUC-UUUUUCUGAAGAGUCGAGUGCCGGUUCGCAAGAACUGGCAUUCAUUUCUUGUAGAAAGAAUAAGAACGGCCAACUGAUGAAUC---UGU-CCAAGU ....(((((...(.((..(((((-(((((((((((((..((((((((((((....))))))))))))..))))).))))))....)))))))..)).)....)))))---...-...... ( -43.70) >consensus ___UAGUUACUUGAUGAUUGUUC_UUUUUCUGAAGAGUCGAGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUGUAAUAACU ...(((((....(.((..(((((.((((((((..(((..((((((((((((....))))))))))))..)))...))))))))....)))))..)).).....)))))............ (-34.00 = -33.45 + -0.55)


| Location | 10,136,679 – 10,136,796 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.40 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -20.55 |
| Energy contribution | -20.42 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.708824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10136679 117 + 20766785 AGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUAGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUUUAACAACUGAAUUGCUUUAAAAUGC---AAUUGUGGGAAGCAAAUAAC (((((((((((....)))))))))))..(((((..(..............(((....))).....................((((((........))---)))))..)))))........ ( -29.70) >DroSec_CAF1 1997 117 + 1 AGUGCCAAGUCGCAAGAUUUGUCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAAUUAAAUGUAAUAACUGAAUUGCUUUCAAAUGC---AAUUGUGCGAAGCAAAUAAC ..(((....(((((.(((((((((((((((((....)))))))))......(.....).))))))))..............((((((........))---)))).))))).)))...... ( -26.40) >DroSim_CAF1 1998 117 + 1 AGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUGUAAUAACUGAAUUGCUUUAAAAUGC---AAUUGUGCGAAGCAAAUAAC ...((((((((....))))))))(((((((((....))))))))).....(((....))).......................(((((((.....((---(....))))))))))..... ( -29.80) >DroEre_CAF1 2017 116 + 1 AGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAACCUAAAUGUAAUGACAUAGUUGAUUUAAAAUGU---AAUUGUUC-AAGCAAAUAAC ...((((((((....))))))))(((((((((....)))))))))..((((((.(((((..........((((....)))))))))..(((.....)---))))))))-........... ( -27.60) >DroYak_CAF1 2006 116 + 1 AGUGCCAAGUCGCAAGAUUUGGCAUUCUAUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUCUAAUAACAAAAUUGAUUUGAAAUGC---AAUUGUGC-AAGCAAAUAAC ...((((((((....))))))))(((((.((((...))))))))).......(((((.((......((((((.(((......))))))))).....)---).))).))-........... ( -27.40) >DroAna_CAF1 2069 113 + 1 AGUGCCGGUUCGCAAGAACUGGCAUUCAUUUCUUGUAGAAAGAAUAAGAACGGCCAACUGAUGAAUC---UGU-CCAAGUGAAAAGCGUUUAAACGCCCCAGUUAUGCA---UUAUUAAA (((((((((((....)))))))))))...(((((((.......)))))))..((.(((((.((....---...-.))........((((....))))..)))))..)).---........ ( -31.00) >consensus AGUGCCAAGUCGCAAGAUUUGGCAUUCUUUUCCUGUGGAAAGAAUAAGAACGGCCAACUGAUAAACUAAAUGUAAUAACUGAAUUGCUUUAAAAUGC___AAUUGUGCGAAGCAAAUAAC ...((((((((....))))))))((((((((......)))))))).....(((....)))..........................................((((.....))))..... (-20.55 = -20.42 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:46 2006