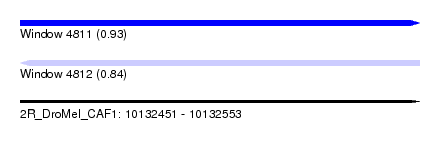
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,132,451 – 10,132,553 |
| Length | 102 |
| Max. P | 0.934139 |

| Location | 10,132,451 – 10,132,553 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -24.65 |
| Energy contribution | -24.98 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.934139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
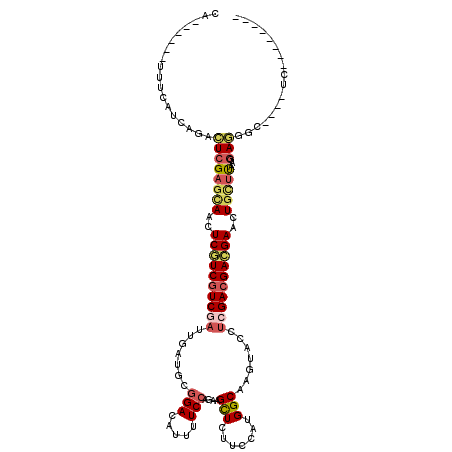
>2R_DroMel_CAF1 10132451 102 + 20766785 --------GA----GCCCUCCUUAAGCAGUUCGUCGUCGAGGUACUUGCCAUGGAAGAGCUUUUGGAAAAUGUCCGCAUCAAUCGACGACGAGUUGCUCGAGUCUGAUGAAA------UG --------..----......((..((((..(((((((((((((....)))......((((....(((.....))))).))..))))))))))..))))..))..........------.. ( -33.80) >DroSec_CAF1 1100 102 + 1 --------GA----GCCCUCCUUAAGCAGUUCGUCGUCGAGGUACUUGCCAUGGAAGAGCUUCUGGAAAAUGUCCGCAUCAAUCGACGACGAGCUGCUCGAGUCUGAUGAAA------UG --------..----......((..((((((((((((((((......(((...((((....))))(((.....))))))....))))))))))))))))..))..........------.. ( -39.90) >DroAna_CAF1 1222 114 + 1 AUAAAUCGCAACUCACUUUCGUUGAAGAGCUCAUCGUCGAUGAUCUUGCCGUGGAACAGCUUCUGGAAAAUGUCGGCAUCAAUGGAGGAGGAGUUGCUCGAGUCUGACGAUA------UU .......(((((((.((((((((((.(((.(((((...))))).)))((((.....(((...)))........)))).))))))))))..)))))))(((.......)))..------.. ( -35.32) >DroSim_CAF1 11682 102 + 1 --------GA----GCCCUCCUUAAGCAGUUCGUCGUCGAGGUACUUGCCAUGGAAGAGCUUCUGGAAAAUGUCCGCAUCAAUCGACGACGAGCUGCUCGAGUCCGAUGAAA------UG --------..----......((..((((((((((((((((......(((...((((....))))(((.....))))))....))))))))))))))))..))..........------.. ( -39.90) >DroWil_CAF1 803 116 + 1 UUAAAUUGGU----ACUUUUUGUAGGCAGCUCAUCGUCAGUAAAUUUGCCAUGGAACAACUUUUGGAAUAUAUCCGCAUCAAUGGACGAUGAAUUAGUCGAAUCCGAUGAAAGACGUCUG ....((((((----((.....)))(((((.((((((((........(((...(......)....(((.....))))))......)))))))).)).)))....)))))............ ( -21.74) >DroYak_CAF1 1424 102 + 1 --------GA----GCCCUCGUUAAGCAGUUCGUCGUCGAGGUACUUGCCAUGAAAAAGCUUCUGGAAAAUGUCCGCAUCAAUCGACGACGAGUUGCUCGAGUCUGAUGAAA------UG --------..----...((((...((((..(((((((((((((....)))..............(((.....))).......))))))))))..))))))))..........------.. ( -33.70) >consensus ________GA____GCCCUCCUUAAGCAGUUCGUCGUCGAGGUACUUGCCAUGGAAGAGCUUCUGGAAAAUGUCCGCAUCAAUCGACGACGAGUUGCUCGAGUCUGAUGAAA______UG .................(((....((((((((((((((((......(((...((((....))))(((.....))))))....)))))))))))))))).))).................. (-24.65 = -24.98 + 0.34)



| Location | 10,132,451 – 10,132,553 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.60 |
| Mean single sequence MFE | -30.27 |
| Consensus MFE | -21.11 |
| Energy contribution | -22.00 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10132451 102 - 20766785 CA------UUUCAUCAGACUCGAGCAACUCGUCGUCGAUUGAUGCGGACAUUUUCCAAAAGCUCUUCCAUGGCAAGUACCUCGACGACGAACUGCUUAAGGAGGGC----UC-------- ..------.....((...((..((((..((((((((((..(.((((((.....)))....(((.......)))..))).)))))))))))..))))..))...)).----..-------- ( -31.10) >DroSec_CAF1 1100 102 - 1 CA------UUUCAUCAGACUCGAGCAGCUCGUCGUCGAUUGAUGCGGACAUUUUCCAGAAGCUCUUCCAUGGCAAGUACCUCGACGACGAACUGCUUAAGGAGGGC----UC-------- ..------.....((...((..(((((.((((((((((..(.((((((.....)))....(((.......)))..))).))))))))))).)))))..))...)).----..-------- ( -34.60) >DroAna_CAF1 1222 114 - 1 AA------UAUCGUCAGACUCGAGCAACUCCUCCUCCAUUGAUGCCGACAUUUUCCAGAAGCUGUUCCACGGCAAGAUCAUCGACGAUGAGCUCUUCAACGAAAGUGAGUUGCGAUUUAU ..------.(((((..(((((((((.................((((((((.(((....))).)))....)))))...(((((...)))))))))....((....)))))))))))).... ( -28.80) >DroSim_CAF1 11682 102 - 1 CA------UUUCAUCGGACUCGAGCAGCUCGUCGUCGAUUGAUGCGGACAUUUUCCAGAAGCUCUUCCAUGGCAAGUACCUCGACGACGAACUGCUUAAGGAGGGC----UC-------- ..------.....((...((..(((((.((((((((((..(.((((((.....)))....(((.......)))..))).))))))))))).)))))..))..))..----..-------- ( -34.60) >DroWil_CAF1 803 116 - 1 CAGACGUCUUUCAUCGGAUUCGACUAAUUCAUCGUCCAUUGAUGCGGAUAUAUUCCAAAAGUUGUUCCAUGGCAAAUUUACUGACGAUGAGCUGCCUACAAAAAGU----ACCAAUUUAA .....((..(((((((((..(((((....(((((.....))))).(((.....)))...))))).)))..((........))...))))))..))...........----.......... ( -20.20) >DroYak_CAF1 1424 102 - 1 CA------UUUCAUCAGACUCGAGCAACUCGUCGUCGAUUGAUGCGGACAUUUUCCAGAAGCUUUUUCAUGGCAAGUACCUCGACGACGAACUGCUUAACGAGGGC----UC-------- ..------..........((((((((..((((((((((..(.((((((.....)))....(((.......)))..))).)))))))))))..))))...))))...----..-------- ( -32.30) >consensus CA______UUUCAUCAGACUCGAGCAACUCGUCGUCGAUUGAUGCGGACAUUUUCCAGAAGCUCUUCCAUGGCAAGUACCUCGACGACGAACUGCUUAAGGAGGGC____UC________ ..................((((((((..((((((((((.......(((.....)))....(((.......))).......))))))))))..)))))...)))................. (-21.11 = -22.00 + 0.89)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:42 2006