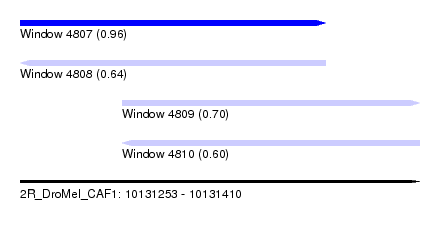
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,131,253 – 10,131,410 |
| Length | 157 |
| Max. P | 0.955570 |

| Location | 10,131,253 – 10,131,373 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -37.73 |
| Energy contribution | -40.07 |
| Covariance contribution | 2.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
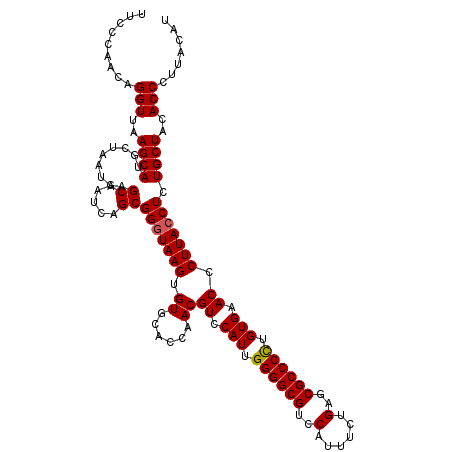
>2R_DroMel_CAF1 10131253 120 + 20766785 UUGUCACUGGCUGCGAAACAAAAAGCAGAAAAAGUUACAAAUGUAAGGGUGUAGCAGAGGUAAGGGUUCACGAGGGCGCUCAGAAAUGGACGCCCCAAUGGACGUUGGUGCACACUUACC ((((.(((..((((..........))))....))).))))..(((((.(((((.(((........(((((...(((((..((....))..)))))...))))).))).))))).))))). ( -41.20) >DroSim_CAF1 10483 120 + 1 UUGUCACUGGCUGCGAAACAAAAAGCAGAAAAAGUUACAAAUGUAAGGGUGUAGCAGAGGUAAGGGUUCACAGGGGCGCUCAGAAAUGGACGCCCCAAUGGACGUUAGUGCACACUUACC ((((.(((..((((..........))))....))).))))..(((((.(((((((....)).((.(((((..((((((..((....))..))))))..))))).))..))))).))))). ( -43.50) >DroYak_CAF1 215 120 + 1 UUGUCACUGGCUGCGAAACAAAAAGCAGAAAGAGUUACAAAUGUAAGGGUGUAGCAGAGUUAAGGGUUCACAGGGGCGCUCCGAAAUGGACGCCCCAAUGGACGUUGGUGCACACUUACC ((((.(((..((((..........))))....))).))))..(((((.(((((.(((........(((((..((((((.(((.....)))))))))..))))).))).))))).))))). ( -42.80) >consensus UUGUCACUGGCUGCGAAACAAAAAGCAGAAAAAGUUACAAAUGUAAGGGUGUAGCAGAGGUAAGGGUUCACAGGGGCGCUCAGAAAUGGACGCCCCAAUGGACGUUGGUGCACACUUACC ((((.(((..((((..........))))....))).))))..(((((.(((((.(((........(((((..((((((..((....))..))))))..))))).))).))))).))))). (-37.73 = -40.07 + 2.33)



| Location | 10,131,253 – 10,131,373 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -32.31 |
| Consensus MFE | -30.93 |
| Energy contribution | -30.71 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10131253 120 - 20766785 GGUAAGUGUGCACCAACGUCCAUUGGGGCGUCCAUUUCUGAGCGCCCUCGUGAACCCUUACCUCUGCUACACCCUUACAUUUGUAACUUUUUCUGCUUUUUGUUUCGCAGCCAGUGACAA ((((((.((......))((.(((.((((((..((....))..)))))).))).)).))))))..................((((.(((....((((..........))))..))).)))) ( -30.80) >DroSim_CAF1 10483 120 - 1 GGUAAGUGUGCACUAACGUCCAUUGGGGCGUCCAUUUCUGAGCGCCCCUGUGAACCCUUACCUCUGCUACACCCUUACAUUUGUAACUUUUUCUGCUUUUUGUUUCGCAGCCAGUGACAA ((((((.((..((....)).(((.((((((..((....))..)))))).))).)).))))))..................((((.(((....((((..........))))..))).)))) ( -33.10) >DroYak_CAF1 215 120 - 1 GGUAAGUGUGCACCAACGUCCAUUGGGGCGUCCAUUUCGGAGCGCCCCUGUGAACCCUUAACUCUGCUACACCCUUACAUUUGUAACUCUUUCUGCUUUUUGUUUCGCAGCCAGUGACAA .(((((((((((.....((.(((.(((((((((.....))).)))))).))).)).........))).))))..))))..((((.(((....((((..........))))..))).)))) ( -33.04) >consensus GGUAAGUGUGCACCAACGUCCAUUGGGGCGUCCAUUUCUGAGCGCCCCUGUGAACCCUUACCUCUGCUACACCCUUACAUUUGUAACUUUUUCUGCUUUUUGUUUCGCAGCCAGUGACAA .(((((((((((.....((.(((.((((((..(......)..)))))).))).)).........))).))))..))))..((((.(((....((((..........))))..))).)))) (-30.93 = -30.71 + -0.22)


| Location | 10,131,293 – 10,131,410 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -43.77 |
| Consensus MFE | -35.33 |
| Energy contribution | -38.33 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.699396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10131293 117 + 20766785 AUGUAAGGGUGUAGCAGAGGUAAGGGUUCACGAGGGCGCUCAGAAAUGGACGCCCCAAUGGACGUUGGUGCACACUUACCCGCUGAUUGCUCAUUAGCAUGCUUAACCUGUUGGGAA ......((((..(((((.((((((.((.(((..(((((..((....))..)))))((((....))))))).)).)))))))((((((.....)))))).))))..))))........ ( -43.80) >DroSim_CAF1 10523 117 + 1 AUGUAAGGGUGUAGCAGAGGUAAGGGUUCACAGGGGCGCUCAGAAAUGGACGCCCCAAUGGACGUUAGUGCACACUUACCCGCUGUUUGCUCAUUAGCAUGCUUAACCUGUUGGAAA ......((((..(((((.((((((.(((((..((((((..((....))..))))))..)))))((....))...)))))))((((.........)))).))))..))))........ ( -44.10) >DroYak_CAF1 255 117 + 1 AUGUAAGGGUGUAGCAGAGUUAAGGGUUCACAGGGGCGCUCCGAAAUGGACGCCCCAAUGGACGUUGGUGCACACUUACCCGCUGAUCGCUCAUUAGAAUGCUUAACCUGUUCGGAA .........((.(((((.((((((.((((...((((((.(((.....)))))))))((((..(((..(((..........)))..).))..)))).)))).)))))))))))))... ( -43.40) >consensus AUGUAAGGGUGUAGCAGAGGUAAGGGUUCACAGGGGCGCUCAGAAAUGGACGCCCCAAUGGACGUUGGUGCACACUUACCCGCUGAUUGCUCAUUAGCAUGCUUAACCUGUUGGGAA ......((((..(((((.((((((.(((((..((((((..((....))..))))))..)))))((....))...)))))))((((((.....)))))).))))..))))........ (-35.33 = -38.33 + 3.00)

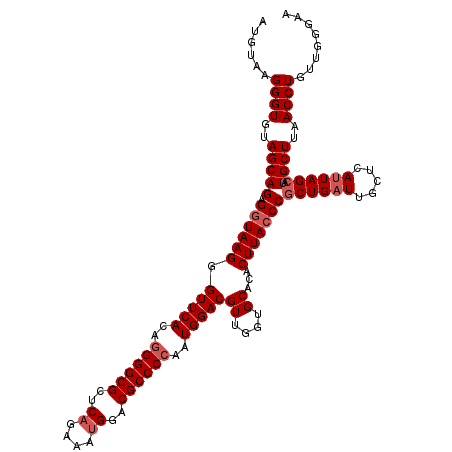
| Location | 10,131,293 – 10,131,410 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -31.75 |
| Energy contribution | -31.87 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10131293 117 - 20766785 UUCCCAACAGGUUAAGCAUGCUAAUGAGCAAUCAGCGGGUAAGUGUGCACCAACGUCCAUUGGGGCGUCCAUUUCUGAGCGCCCUCGUGAACCCUUACCUCUGCUACACCCUUACAU .........(((..((((((((....))))......(((((((.((......))((.(((.((((((..((....))..)))))).))).)).))))))).))))..)))....... ( -34.80) >DroSim_CAF1 10523 117 - 1 UUUCCAACAGGUUAAGCAUGCUAAUGAGCAAACAGCGGGUAAGUGUGCACUAACGUCCAUUGGGGCGUCCAUUUCUGAGCGCCCCUGUGAACCCUUACCUCUGCUACACCCUUACAU .........(((..((((((((....))))......(((((((.((..((....)).(((.((((((..((....))..)))))).))).)).))))))).))))..)))....... ( -37.10) >DroYak_CAF1 255 117 - 1 UUCCGAACAGGUUAAGCAUUCUAAUGAGCGAUCAGCGGGUAAGUGUGCACCAACGUCCAUUGGGGCGUCCAUUUCGGAGCGCCCCUGUGAACCCUUAACUCUGCUACACCCUUACAU .......((((((((((((.((.((..((.....))..)).)).))))......((.(((.(((((((((.....))).)))))).))).))..))))).))).............. ( -35.10) >consensus UUCCCAACAGGUUAAGCAUGCUAAUGAGCAAUCAGCGGGUAAGUGUGCACCAACGUCCAUUGGGGCGUCCAUUUCUGAGCGCCCCUGUGAACCCUUACCUCUGCUACACCCUUACAU .........(((..((((.........((.....))(((((((.((......))((.(((.((((((..(......)..)))))).))).)).))))))).))))..)))....... (-31.75 = -31.87 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:40 2006