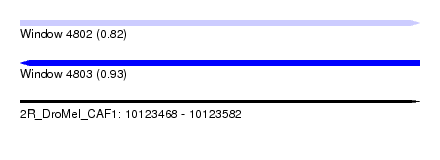
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,123,468 – 10,123,582 |
| Length | 114 |
| Max. P | 0.929129 |

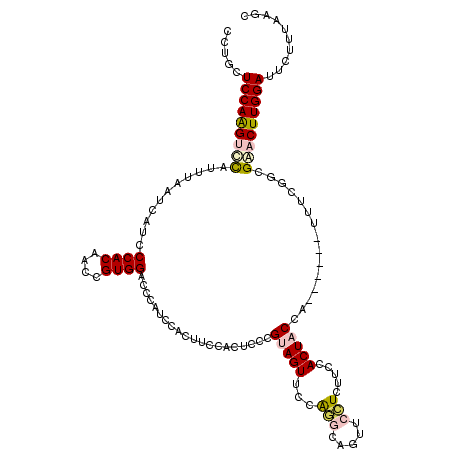
| Location | 10,123,468 – 10,123,582 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -22.62 |
| Energy contribution | -24.46 |
| Covariance contribution | 1.84 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10123468 114 + 20766785 GCUUAAAGAAUCCAAGUCCGCCGAAA------UGGUAGUGGAAGAGGAACUGCCUGAAACUACGGGAGUGGAAGUAGAUGGGUCCACGGUUGUGGGAUGAUUAAAUAGACCUGGAUCAGG ..........((((..((((......------.((((((.........))))))........))))..))))........((((((.((((.(((.....)))....))))))))))... ( -30.46) >DroSec_CAF1 7271 114 + 1 GCUUAAAGAAUCCAAGGUCGCCGAAA------UGGUAGUGGAAGAGGAACUGCCUGGAACUACGGGAGUGGAAGUGGACGGGUCCACGGUUGUGGGAUGAUUAAAUGGACUUGGAGCAGG ((((.(((..((((.(((((((....------.((((((.........)))))).....((((....))))..(((((....))))).......)).)))))...))))))).))))... ( -34.70) >DroSim_CAF1 7072 114 + 1 GCUUAAAGAAUCCAAGGUCGCCGAAU------UGGUAGUGGAAGAGGAACUGCCUGGAACUACGGGAGUGGAAGUGGACGGGUCCACGGUUGUGGGAUGAUUAAAUGGACUUGGAGCAGG ((((.(((..((((.(((((((....------.((((((.........)))))).....((((....))))..(((((....))))).......)).)))))...))))))).))))... ( -34.70) >DroEre_CAF1 7461 105 + 1 GCUUAGGGAAUCCAAGUUCGCCGAAAUUAGCAUUGAAGUGGGAGACGAGCUAGGUGGAACUACGGG---------------AUCCACGCUUGUGGGAAUAUUAAAGGAACCUGGAGCAGG ((((.(((..(((..((((.(((.....(((.....(((.(....)..)))..(((((.(....).---------------.))))))))..)))))))......))).))).))))... ( -30.10) >DroYak_CAF1 7381 120 + 1 GCUAAGGGAAUCCAAGUUCGCCGAAAUCAGCAUGGAAGUGGGAGAGAGACUUGGCGGAACUACGGGAGUGGAAGUGGAUGGACCCACGCAUGUGGGAAUAUUAAAGGAACUUGGAGCCGG .....((...((((((((((((((..((..(((....))).)).......)))))....((((....)))).....((((..(((((....)))))..))))....))))))))).)).. ( -36.90) >consensus GCUUAAAGAAUCCAAGUUCGCCGAAA______UGGUAGUGGAAGAGGAACUGCCUGGAACUACGGGAGUGGAAGUGGACGGGUCCACGGUUGUGGGAUGAUUAAAUGGACUUGGAGCAGG ..........(((((((((.(((..........((((((.........)))))).....((((....)))).......))).(((((....)))))..........)))))))))..... (-22.62 = -24.46 + 1.84)

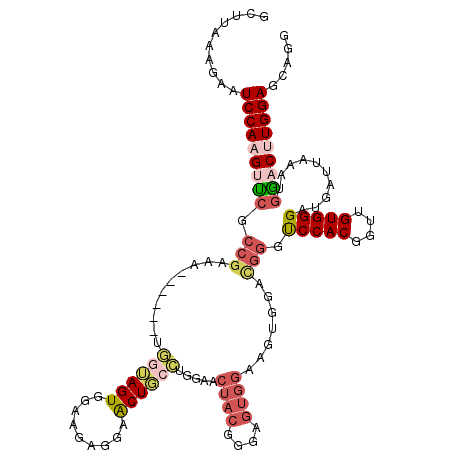
| Location | 10,123,468 – 10,123,582 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -23.74 |
| Consensus MFE | -16.66 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10123468 114 - 20766785 CCUGAUCCAGGUCUAUUUAAUCAUCCCACAACCGUGGACCCAUCUACUUCCACUCCCGUAGUUUCAGGCAGUUCCUCUUCCACUACCA------UUUCGGCGGACUUGGAUUCUUUAAGC ...(((((((((((...................(((((..........)))))....(((((....((.((.....)).)))))))..------.......)))))))))))........ ( -25.50) >DroSec_CAF1 7271 114 - 1 CCUGCUCCAAGUCCAUUUAAUCAUCCCACAACCGUGGACCCGUCCACUUCCACUCCCGUAGUUCCAGGCAGUUCCUCUUCCACUACCA------UUUCGGCGACCUUGGAUUCUUUAAGC .....(((((((((...................(((((....)))))..........(((((....((.((.....)).)))))))..------.....).)).)))))).......... ( -20.80) >DroSim_CAF1 7072 114 - 1 CCUGCUCCAAGUCCAUUUAAUCAUCCCACAACCGUGGACCCGUCCACUUCCACUCCCGUAGUUCCAGGCAGUUCCUCUUCCACUACCA------AUUCGGCGACCUUGGAUUCUUUAAGC .....(((((((((...................(((((....)))))..........(((((....((.((.....)).)))))))..------.....).)).)))))).......... ( -20.80) >DroEre_CAF1 7461 105 - 1 CCUGCUCCAGGUUCCUUUAAUAUUCCCACAAGCGUGGAU---------------CCCGUAGUUCCACCUAGCUCGUCUCCCACUUCAAUGCUAAUUUCGGCGAACUUGGAUUCCCUAAGC .....(((((((((.............((.((((((((.---------------........)))))...))).)).............(((......)))))))))))).......... ( -22.80) >DroYak_CAF1 7381 120 - 1 CCGGCUCCAAGUUCCUUUAAUAUUCCCACAUGCGUGGGUCCAUCCACUUCCACUCCCGUAGUUCCGCCAAGUCUCUCUCCCACUUCCAUGCUGAUUUCGGCGAACUUGGAUUCCCUUAGC ..((.(((((((((..........(((((....)))))...........................((((((((...................))))).))))))))))))..))...... ( -28.81) >consensus CCUGCUCCAAGUCCAUUUAAUCAUCCCACAACCGUGGACCCAUCCACUUCCACUCCCGUAGUUCCAGGCAGUUCCUCUUCCACUACCA______UUUCGGCGAACUUGGAUUCUUUAAGC .....(((((((((...........((((....))))....................(((((...(((.....))).....)))))...............))))))))).......... (-16.66 = -17.50 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:33 2006