| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,114,771 – 10,114,891 |
| Length | 120 |
| Max. P | 0.608954 |

| Location | 10,114,771 – 10,114,891 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
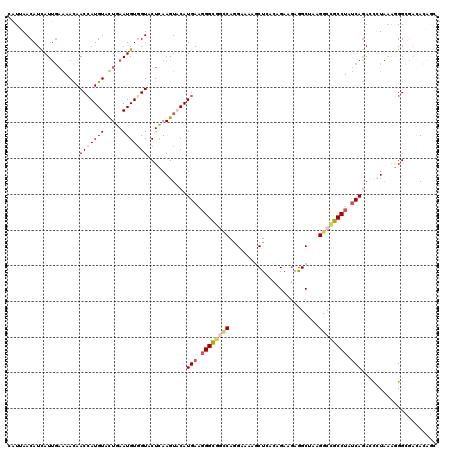
| Mean pairwise identity | 77.83 |
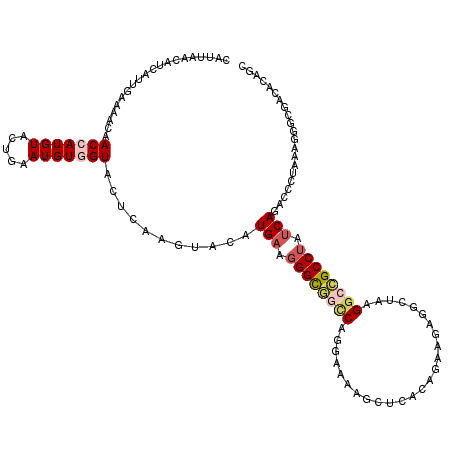
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -16.48 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608954 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 10114771 120 - 20766785 CAUAAACAUCAUUGAAAACAACCAUGUAUUGAAUGUGGUACUCAAGUACAUGAAGGGCGGCCAGGAAAAGCUCACAGAAGAGGCUAAGGCCGCCUAUCAGACCCUAAAGGGUGAAACAGC ......((.((((.((.(((....))).)).))))))((((....)))).(((.((((((((......(((((......)).)))..)))))))).))).((((....))))........ ( -35.90) >DroGri_CAF1 4503 120 - 1 UGUGAACAUCAUUGAGAAUAACAAUGUACUCAAUGUGGUACUGAAGUUCAUGAAGGGUCAGCAGGAAAAUCUAAGUGAGCAGGCCAGGGCUGCCUACCAAAAUAUGCGUGGCAGCAACAG ......((.(((((((.(((....))).)))))))))...(((..((((((..(((.((.....))...)))..))))))........((((((.((((.....)).))))))))..))) ( -32.60) >DroSim_CAF1 5503 120 - 1 CCUUAACAUCAUUGAAAACAACCAUGUAUUGAAUGUGGUACUCAAGUACAUGAAGGGCGGCCAGGAGAAGCUCACAGAAGAGGCGAAGGCCGCCUAUCAGACCAUAAAGGGUGAAACAGC ((((..((.((((.((.(((....))).)).))))))((((....)))).(((.((((((((....(...(((......))).)...)))))))).))).......)))).......... ( -33.60) >DroEre_CAF1 4365 120 - 1 CAUUAACAUCAUUGAAAACAACCAUGUACUGAAUGUCGUUCUCAAGUACAUGAAGGGUGGCCAGGAGAAGCUCACAGACGAGGCUAAGGCCGCCUAUCAGACCCUAAAGGGCGACACAGC .....((((..(((....)))..)))).(((..(((((((((..((....(((.((((((((...((...(((......))).))..)))))))).)))....))..)))))))))))). ( -38.60) >DroWil_CAF1 4432 120 - 1 UGUCAACAUUAUCCUGAAUAACCAUGUUCUAAAUGUGGUUCUCAAGUUCAUGAACGGCAAUCAAGAAAAACUAAGCGAGGAGGCGAUGGCUGCCUAUCAGAAUCUGAAAGGAUCCAACUC ..........(((((....((((((((.....)))))))).(((..(((.(((..((((..((((.....))..((......))..))..))))..))))))..))).)))))....... ( -25.50) >DroYak_CAF1 4246 120 - 1 CAUUAACAUCAUUGAAAACAACCAUGUAUUGAAUGUGGUUCUUAAGUACAUGAAGGGCGGCCAAGAGAAGCUGACAGACGAAGCUAAGGCCGCCUAUCAGACCCUAAAAGGCGACACAGC .....((((..(((....)))..))))......(((.((..((.((....(((.((((((((......((((.........))))..)))))))).)))....)).))..)).))).... ( -31.20) >consensus CAUUAACAUCAUUGAAAACAACCAUGUACUGAAUGUGGUACUCAAGUACAUGAAGGGCGGCCAGGAAAAGCUCACAGAAGAGGCUAAGGCCGCCUAUCAGACCCUAAAGGGCGACACAGC ....................(((((((.....)))))))...........(((.((((((((.........................)))))))).)))..................... (-16.48 = -17.26 + 0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:27 2006