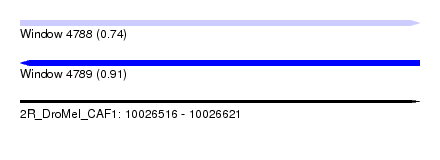
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 10,026,516 – 10,026,621 |
| Length | 105 |
| Max. P | 0.907779 |

| Location | 10,026,516 – 10,026,621 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -17.43 |
| Energy contribution | -17.60 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
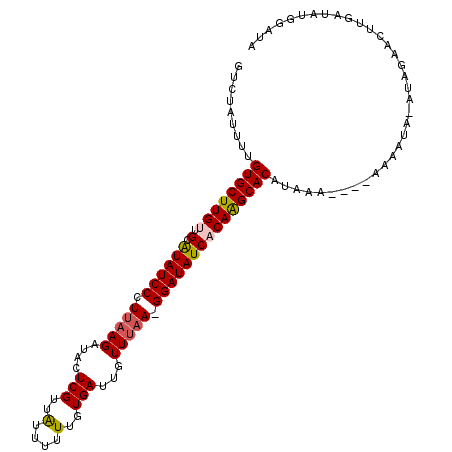
>2R_DroMel_CAF1 10026516 105 + 20766785 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUAA-GGAUAUCACAAGCACGUAAA----AAAAUA-AUAGAACUUGAUAUGGAUA .((((((.(((((((((.(.((((((.(((((......(((((.....)))))..)))))-))))))))))))))))....----.....)-))))).............. ( -23.00) >DroSec_CAF1 5133 104 + 1 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUAA-GGAUAUCUCAAGCACGUAAA-----AAAUA-AUAGAACUUGAUAUGGAUA .((((((.((((((((..(.((((((.(((((......(((((.....)))))..)))))-))))))).))))))))....-----....)-))))).............. ( -21.40) >DroEre_CAF1 5114 105 + 1 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUAA-GGAUAUCACAAGCACUUACA----AAAAUA-AUAGAACUUGAUAUGGAUA .((((((..((((((((.(.((((((.(((((......(((((.....)))))..)))))-))))))))))))))).....----.....)-))))).............. ( -22.10) >DroYak_CAF1 5119 109 + 1 GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUAA-GGAUAUCACAAGCACUAAAAAAAAAAAAUA-AUAGAACUUGAUAUGGAUA .((((((..((((((((.(.((((((.(((((......(((((.....)))))..)))))-)))))))))))))))..............)-))))).............. ( -21.79) >DroAna_CAF1 4866 100 + 1 GUGUAUUUUGUGCUUGUUGCGUAUCCCUUGAGAUACUCGUUGAUUUUUGUGAUUUUUGAAUGGAUAUCACAGGCACACCACUCU-AAAGCAAGCAACACU----------C ((((......(((((((((.(((((......))))).).......((((((((.(((....))).))))))))...........-..)))))))))))).----------. ( -22.80) >DroPer_CAF1 5119 88 + 1 GUGUAUUUUGUGCUUGUUGCAUAUCCCUAAAGAUACUCGUUGUU-UUUGUGAUUGUUUAA-GGAUAUCUCAAGCACACUAA----AAAAUA-AAA---------------- ........((((((((..(.((((((.((((.(....((.....-..))....).)))).-))))))).))))))))....----......-...---------------- ( -19.10) >consensus GUCUAUUUUGUGCUUGUUGCAUAUCCCUUAAGAUACUCGUUAUUUUUUGUGAUUGUUUAA_GGAUAUCACAAGCACAUAAA____AAAAUA_AUAGAACUUGAUAUGGAUA .........((((((((.(.((((((.(((((....(((..(....)..)))...))))).)))))))))))))))................................... (-17.43 = -17.60 + 0.17)


| Location | 10,026,516 – 10,026,621 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -19.84 |
| Consensus MFE | -15.82 |
| Energy contribution | -16.82 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
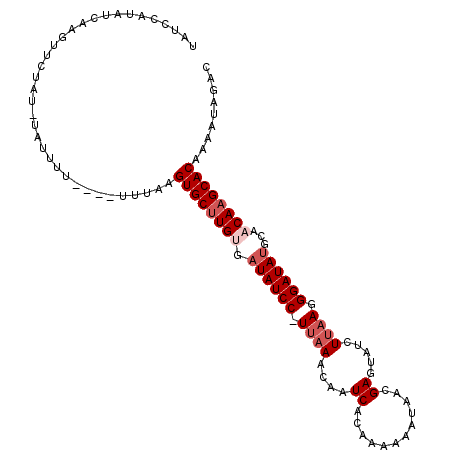
>2R_DroMel_CAF1 10026516 105 - 20766785 UAUCCAUAUCAAGUUCUAU-UAUUUU----UUUACGUGCUUGUGAUAUCC-UUAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ...................-......----.....((((((((.((((((-((......((............))........))))))))...))))))))......... ( -18.94) >DroSec_CAF1 5133 104 - 1 UAUCCAUAUCAAGUUCUAU-UAUUU-----UUUACGUGCUUGAGAUAUCC-UUAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ..................(-(((((-----(....(((((((..((((((-((......((............))........))))))))....)))))))))))))).. ( -16.24) >DroEre_CAF1 5114 105 - 1 UAUCCAUAUCAAGUUCUAU-UAUUUU----UGUAAGUGCUUGUGAUAUCC-UUAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ..............(((((-(((...----.))))((((((((.((((((-((......((............))........))))))))...))))))))....)))). ( -20.04) >DroYak_CAF1 5119 109 - 1 UAUCCAUAUCAAGUUCUAU-UAUUUUUUUUUUUUAGUGCUUGUGAUAUCC-UUAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ...................-...............((((((((.((((((-((......((............))........))))))))...))))))))......... ( -18.84) >DroAna_CAF1 4866 100 - 1 G----------AGUGUUGCUUGCUUU-AGAGUGGUGUGCCUGUGAUAUCCAUUCAAAAAUCACAAAAAUCAACGAGUAUCUCAAGGGAUACGCAACAAGCACAAAAUACAC .----------.(((((..((((...-.(((((((((.......))).)))))).....................((((((....))))))))))..)))))......... ( -24.60) >DroPer_CAF1 5119 88 - 1 ----------------UUU-UAUUUU----UUAGUGUGCUUGAGAUAUCC-UUAAACAAUCACAAA-AACAACGAGUAUCUUUAGGGAUAUGCAACAAGCACAAAAUACAC ----------------...-......----....((((((((..((((((-(((((...((.....-......)).....)))))))))))....))))))))........ ( -20.40) >consensus UAUCCAUAUCAAGUUCUAU_UAUUUU____UUUAAGUGCUUGUGAUAUCC_UUAAACAAUCACAAAAAAUAACGAGUAUCUUAAGGGAUAUGCAACAAGCACAAAAUAGAC ...................................((((((((.((((((.((((....((............)).....)))).))))))...))))))))......... (-15.82 = -16.82 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:19 2006