| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,198,697 – 1,198,797 |
| Length | 100 |
| Max. P | 0.857229 |

| Location | 1,198,697 – 1,198,797 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.04 |
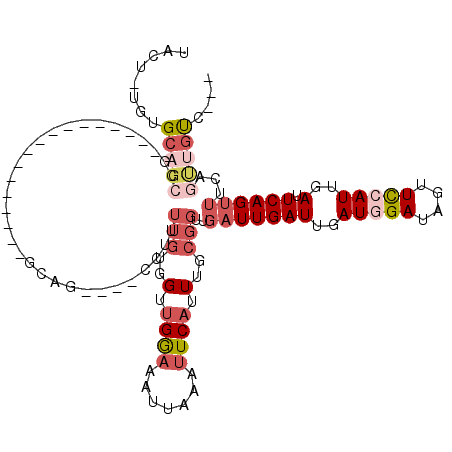
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -10.98 |
| Energy contribution | -12.78 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857229 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
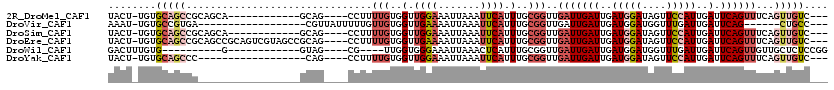
>2R_DroMel_CAF1 1198697 100 - 20766785 UACU-UGUGCAGCCGCAGCA------------GCAG----CCUUUUGUGGUUGGAAAUUAAAUUCAUUUGCGGUUGAUUGAUUGAUGGAUAGUUCCAUUGAUUCAGUUUCAGUUGUC--- ....-....(((((((((..------------.(((----((......)))))(((......)))..)))))))))((((((..(((((....)))))..).)))))..........--- ( -31.50) >DroVir_CAF1 18028 92 - 1 AAAU-UGUGCCGUGA------------------CGUUAUUUUUGUUGUGGUUGAAAAUUAAAUUCAUUUGCGGUUGAUUGAUUGAUGGAUGGUUUGAUUGAUUCAG------CUGCC--- .(((-(..((((..(------------------((.......)))..))))....))))..........(((((((((..((..(........)..))..).))))------)))).--- ( -24.30) >DroSim_CAF1 13522 100 - 1 UACU-UGUGCAGCCGCAGCA------------GCAG----CCUUUUGUGGUUGGAAAUUAAAUUCAUUUGCGGUUGAUUGAUUGAUGGAUAGUUCCAUUGAUUCAGUUUCAGUUGUC--- ....-....(((((((((..------------.(((----((......)))))(((......)))..)))))))))((((((..(((((....)))))..).)))))..........--- ( -31.50) >DroEre_CAF1 17247 112 - 1 UACU-UGUGCAGCCGCAGCCGCAGUCGUAGCCGCAG----CCUUUUGUGGUUGAAAAUUAAAUUCAUUUGCGGUUGAUUGAUUGAUGGAUAGUUCCAUUGAUUCAGUUUCAGUUGUC--- ....-...(((((..(((((((((....((((((((----....))))))))(((.......)))..)))))))))((((((..(((((....)))))..).)))))....))))).--- ( -36.40) >DroWil_CAF1 23492 90 - 1 GACUUUGUG----------G------------GUAG----CG----UUGGUGGGAAAUUAAACUCAUUUGCGGUUGAUUGAUUGAUGGAUGGUUUGAUUGAUUCAGUUGUUGCUCUCCGG .......((----------(------------..((----((----..(.((((.((((((((.(((..((((((....))))).)..)))))))))))..)))).)...))))..))). ( -22.70) >DroYak_CAF1 11795 94 - 1 UACU-UGUGCAGCCC------------------CAG----CCUUUUGUGGUUGGAAAUUAAAUUCAUUUGCGGUUGAUUGAUUGAUGGAUAGUUCCAUUGAUUCAGUUUCAGUUGUC--- ....-...(((((.(------------------(((----((......)))))).....................(((((((..(((((....)))))..).))))))...))))).--- ( -25.90) >consensus UACU_UGUGCAGCCG_________________GCAG____CCUUUUGUGGUUGGAAAUUAAAUUCAUUUGCGGUUGAUUGAUUGAUGGAUAGUUCCAUUGAUUCAGUUUCAGUUGUC___ ........(((((...............................(((..(.((((.......)))).)..)))..(((((((..(((((....)))))..).))))))...))))).... (-10.98 = -12.78 + 1.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:53 2006