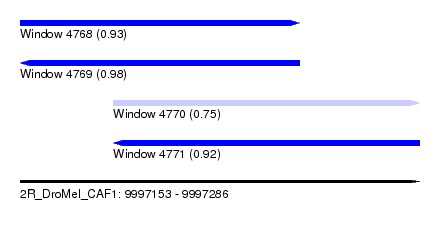
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,997,153 – 9,997,286 |
| Length | 133 |
| Max. P | 0.983638 |

| Location | 9,997,153 – 9,997,246 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933699 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9997153 93 + 20766785 UUCUUUAUGUUUUGUGUGCCGGCUUUAUUUU--------UUU--------UGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC--ACACAAAACAAACAUAGGCCUAUACA .......(((((((((((.((((........--------...--------.))))........((((...........)))))--))))))))))................ ( -20.30) >DroSec_CAF1 731 100 + 1 UUCUUUAUGUUUUGUGUGCCGGCUUUAUUUU--------UUUUUGCUUUCUGCCGUUUCUGCCUGUG---CCUAUUCUCACACACACACAAAACAAACAUAGGCCUAUACA .......(((((((((((.((((........--------............))))........((((---........))))..)))))))))))................ ( -20.75) >DroEre_CAF1 667 105 + 1 UUCUUUAUGUUUUGUGCGCCGGCCUA-U---GGCUUUUGCUUUUGCUUUUUGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC--ACACAAAACAAACAUAGGCCUAUGCA ..............((((..((((((-(---(((....((....)).....)))((((.((..((((.............)))--)..))))))....)))))))..)))) ( -26.22) >DroYak_CAF1 726 103 + 1 UUCUUUAUGUU--GUGUGCCGGCCUA-U---GGCUUUUGCUUUUGCUUUUUGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC--ACACAAAACAAACAUAGGCCCAUACA ..........(--(((((..((((((-(---(((....((....)).....)))((((.((..((((.............)))--)..))))))....))))))))))))) ( -27.12) >consensus UUCUUUAUGUUUUGUGUGCCGGCCUA_U___________CUUUUGCUUUUUGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC__ACACAAAACAAACAUAGGCCUAUACA .............(((((.((((............................)))).....(((((((..............................))))))).))))). (-13.52 = -13.40 + -0.12)

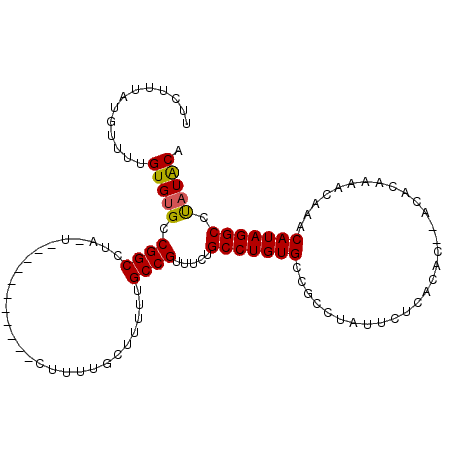
| Location | 9,997,153 – 9,997,246 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -27.69 |
| Consensus MFE | -17.09 |
| Energy contribution | -16.96 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9997153 93 - 20766785 UGUAUAGGCCUAUGUUUGUUUUGUGU--GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCA--------AAA--------AAAAUAAAGCCGGCACACAAAACAUAAAGAA ................((((((((((--(((((.(......)..))).........((((.--------...--------........))))))))))))))))....... ( -23.70) >DroSec_CAF1 731 100 - 1 UGUAUAGGCCUAUGUUUGUUUUGUGUGUGUGUGAGAAUAGG---CACAGGCAGAAACGGCAGAAAGCAAAAA--------AAAAUAAAGCCGGCACACAAAACAUAAAGAA ................((((((((((((((((........)---))).........((((............--------........))))))))))))))))....... ( -24.85) >DroEre_CAF1 667 105 - 1 UGCAUAGGCCUAUGUUUGUUUUGUGU--GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCAAAAAGCAAAAGCAAAAGCC---A-UAGGCCGGCGCACAAAACAUAAAGAA .((...(((((((.....((((((.(--((((..(......).))))).))))))..(((.....((....))....)))---)-)))))).))................. ( -31.60) >DroYak_CAF1 726 103 - 1 UGUAUGGGCCUAUGUUUGUUUUGUGU--GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCAAAAAGCAAAAGCAAAAGCC---A-UAGGCCGGCACAC--AACAUAAAGAA (((.(.(((((((.....((((((.(--((((..(......).))))).))))))..(((.....((....))....)))---)-)))))).).))).--........... ( -30.60) >consensus UGUAUAGGCCUAUGUUUGUUUUGUGU__GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCAAAAAGCAAAAA___________A_AAAGCCGGCACACAAAACAUAAAGAA ...((((((....))))))(((((((..(((((.......((.......)).....((((............................)))).)))))...)))))))... (-17.09 = -16.96 + -0.13)

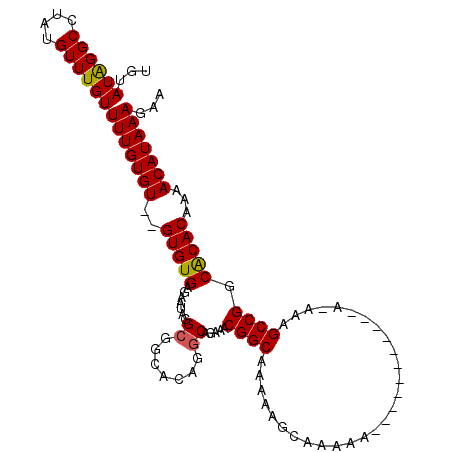
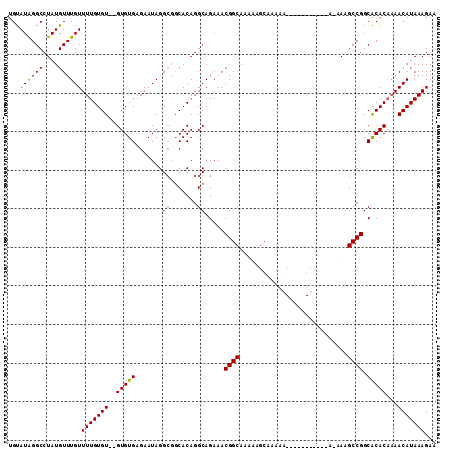
| Location | 9,997,184 – 9,997,286 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -18.68 |
| Energy contribution | -19.06 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9997184 102 + 20766785 UUU--------UGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC--ACACAAAACAAACAUAGGCCUAUACAUACAUAUGUGUGUGCAUAUGUAGGCCUGGAAUCGUGAAUU .((--------..(.(((((.(((((((..((((((........--.............))))))...))))(((((((((....))))))))))))..))))).)..)).. ( -27.20) >DroSec_CAF1 762 109 + 1 UUUUUGCUUUCUGCCGUUUCUGCCUGUG---CCUAUUCUCACACACACACAAAACAAACAUAGGCCUAUACAUACAUAUGUAUGUGCACAUGUAGGCCUGGAAUCGGGAAUU .............(((.(((((..((((---........))))..)).............(((((((((((((((....))))))......)))))))))))).)))..... ( -26.20) >DroSim_CAF1 768 112 + 1 UUUUUGCUUUCUGCCGUUUCUGCCUGUGCCGCCUAUUCUCACACACACACAAAACAAACAUAGGCCUAUACAUACAUAUGUAUGUGCACAUGUAGGCCUGGAAUCGUGAAUU ..((..(......(((..(((((.(((((.((((((.......................))))))....((((((....))))))))))).)))))..)))....)..)).. ( -27.70) >DroEre_CAF1 702 102 + 1 CUUUUGCUUUUUGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC--ACACAAAACAAACAUAGGCCUAUGCAUA--------UGUACAUAUGUAGGCCUGGAACCGGGAAUA ..........((.(((.(((((..((((.............)))--)..)).........(((((((..(((((--------(....)))))))))))))))).))).)).. ( -24.02) >DroYak_CAF1 759 102 + 1 CUUUUGCUUUUUGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC--ACACAAAACAAACAUAGGCCCAUACAUA--------UGUACAUAUGUAGGCCUGGAAUCGGGAAUA ..........((.(((.(((((..((((.............)))--)..)).........((((((..((((((--------(....)))))))))))))))).))).)).. ( -23.52) >consensus UUUUUGCUUUCUGCCGUUUCUGCCUGUGCCGCCUAUUCUCACAC__ACACAAAACAAACAUAGGCCUAUACAUACAUAUGU_UGUGCAUAUGUAGGCCUGGAAUCGGGAAUU .............(((..(((((.((((..((((((.......................))))))...(((((((....))))))))))).)))))..)))........... (-18.68 = -19.06 + 0.38)

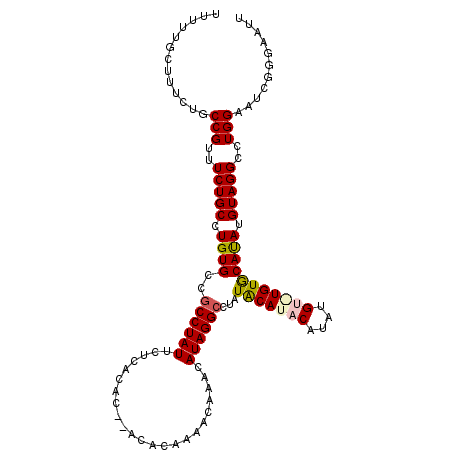
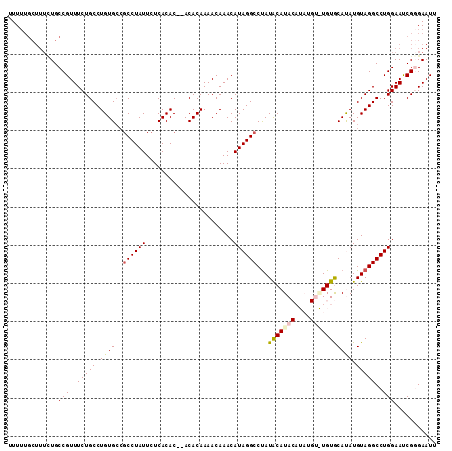
| Location | 9,997,184 – 9,997,286 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 86.26 |
| Mean single sequence MFE | -32.30 |
| Consensus MFE | -23.30 |
| Energy contribution | -24.90 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9997184 102 - 20766785 AAUUCACGAUUCCAGGCCUACAUAUGCACACACAUAUGUAUGUAUAGGCCUAUGUUUGUUUUGUGU--GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCA--------AAA ......((.(((.(((((((.(((((((........))))))).))))))).(((((((.((((.(--((......))).)))).)))))))))).))...--------... ( -32.80) >DroSec_CAF1 762 109 - 1 AAUUCCCGAUUCCAGGCCUACAUGUGCACAUACAUAUGUAUGUAUAGGCCUAUGUUUGUUUUGUGUGUGUGUGAGAAUAGG---CACAGGCAGAAACGGCAGAAAGCAAAAA ..((((((.....(((((((.(((((((........))))))).))))))).......((((((.(((((.((....)).)---)))).)))))).)))..)))........ ( -33.50) >DroSim_CAF1 768 112 - 1 AAUUCACGAUUCCAGGCCUACAUGUGCACAUACAUAUGUAUGUAUAGGCCUAUGUUUGUUUUGUGUGUGUGUGAGAAUAGGCGGCACAGGCAGAAACGGCAGAAAGCAAAAA ..(((........(((((((.(((((((........))))))).))))))).((((..((((((.(((((((........)).))))).))))))..)))))))........ ( -32.80) >DroEre_CAF1 702 102 - 1 UAUUCCCGGUUCCAGGCCUACAUAUGUACA--------UAUGCAUAGGCCUAUGUUUGUUUUGUGU--GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCAAAAAGCAAAAG .....(((.(((.((((((((((((....)--------))))..))))))).(((((((.((((.(--((......))).)))).)))))))))).)))............. ( -31.00) >DroYak_CAF1 759 102 - 1 UAUUCCCGAUUCCAGGCCUACAUAUGUACA--------UAUGUAUGGGCCUAUGUUUGUUUUGUGU--GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCAAAAAGCAAAAG .....(((.(((.((((((((((((....)--------)))))..)))))).(((((((.((((.(--((......))).)))).)))))))))).)))............. ( -31.40) >consensus AAUUCCCGAUUCCAGGCCUACAUAUGCACA_ACAUAUGUAUGUAUAGGCCUAUGUUUGUUUUGUGU__GUGUGAGAAUAGGCGGCACAGGCAGAAACGGCAAAAAGCAAAAA .....(((.(((.(((((((.(((((((........))))))).))))))).(((((((.((((.(..((......))).)))).)))))))))).)))............. (-23.30 = -24.90 + 1.60)

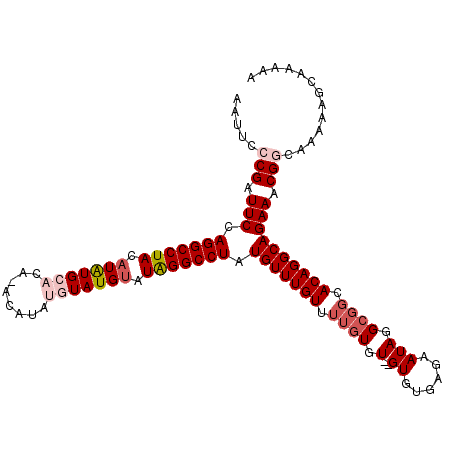
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:42:01 2006