| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,698,456 – 1,698,546 |
| Length | 90 |
| Max. P | 0.686664 |

| Location | 1,698,456 – 1,698,546 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -22.63 |
| Consensus MFE | -15.64 |
| Energy contribution | -16.17 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686664 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
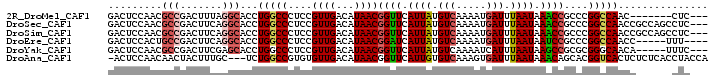
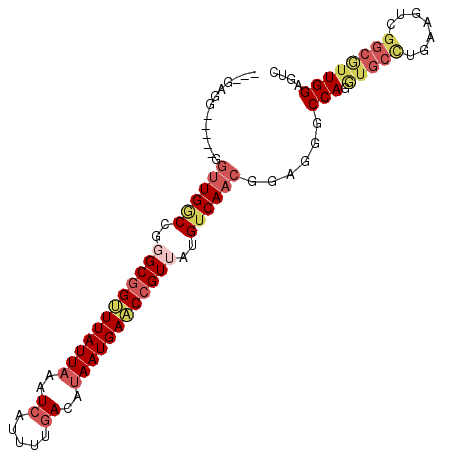
>2R_DroMel_CAF1 1698456 90 + 20766785 GACUCCAACGCCGACUUUAGGCACCUGGCCCUCCGUUGACAUAACGGUUCAUUAUGUCAAAAUGAUUUAAUAAACCGCCCGGCCAAC-------CUC--- .........(((.......)))...(((((....((((...))))((((.((((.(((.....))).)))).))))....)))))..-------...--- ( -22.20) >DroSec_CAF1 84283 97 + 1 GACUCCAACGCCGACUUCAGGCACCUGGCCCUCCGUUGACAUAACGGUUCAUUAUGUCAAAAUGAUUUAAUAAACCGCCCGGCCAACCGCCAGCCUC--- .........(((.......)))..(((((.....((((.(....(((((.((((.(((.....))).)))).)))))....).)))).)))))....--- ( -24.90) >DroSim_CAF1 81760 97 + 1 GACUCCAACGCCGACUUCAGGCACCUGGCCCUCCGUUGACAUAACGGUUCAUUAUGUCAAAAUGAUUUAAUAAACCGCCCGGCCAACCGCCAGCCUC--- .........(((.......)))..(((((.....((((.(....(((((.((((.(((.....))).)))).)))))....).)))).)))))....--- ( -24.90) >DroEre_CAF1 89748 91 + 1 GACUCCACUGCCGACUUCAGGCACCUGGCCCUCCGUUGACAUAACGGAUCAUUAUGUCAAAAUGAUUUAAUAAUCCGCCCGGCCAACC-----UUU---- ........((((.......))))..(((((....((((...))))((((.((((.(((.....))).)))).))))....)))))...-----...---- ( -22.80) >DroYak_CAF1 9891 92 + 1 GACUCCAACGCCGACUUCGAGCACCUGGCCCUCCGUUGACAUAACGGUUCAUUAUGUCAAAAUCAUUUAAUAAGCCGCGCGGGCAACA-----UUUC--- .........((((....)).)).....((((....(((((((((.......))))))))).............((...))))))....-----....--- ( -19.70) >DroAna_CAF1 10815 96 + 1 -ACUCCAACAACUACUUUGC---UCUGGCCGUGUGUUGACAUAACGGUUCAUUGUGUCAAAGUGAUUUAAUAAACAGCACGGUCACUCUCUCACCUACCA -...................---..(((((((((.(((((((((.......))))))))).((..........)).)))))))))............... ( -21.30) >consensus GACUCCAACGCCGACUUCAGGCACCUGGCCCUCCGUUGACAUAACGGUUCAUUAUGUCAAAAUGAUUUAAUAAACCGCCCGGCCAACC_____CCUC___ .........(((.......)))...(((((....((((...))))((((.((((.(((.....))).)))).))))....)))))............... (-15.64 = -16.17 + 0.53)
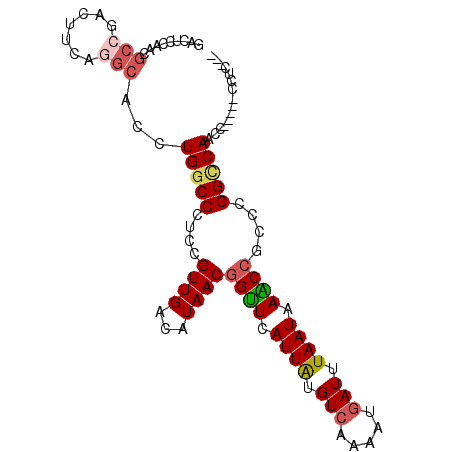
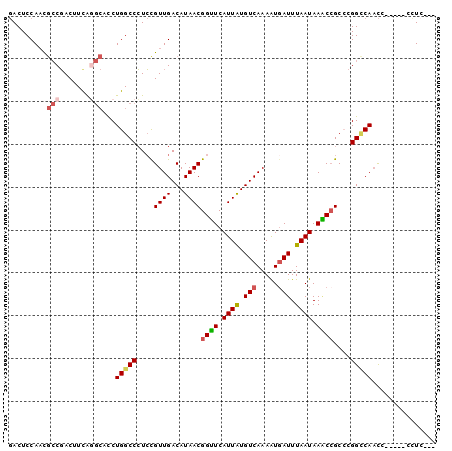
| Location | 1,698,456 – 1,698,546 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -20.48 |
| Energy contribution | -22.15 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.519596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1698456 90 - 20766785 ---GAG-------GUUGGCCGGGCGGUUUAUUAAAUCAUUUUGACAUAAUGAACCGUUAUGUCAACGGAGGGCCAGGUGCCUAAAGUCGGCGUUGGAGUC ---...-------((((((..((((((((((((..((.....))..))))))))))))..))))))......(((...(((.......)))..))).... ( -28.30) >DroSec_CAF1 84283 97 - 1 ---GAGGCUGGCGGUUGGCCGGGCGGUUUAUUAAAUCAUUUUGACAUAAUGAACCGUUAUGUCAACGGAGGGCCAGGUGCCUGAAGUCGGCGUUGGAGUC ---((.((((((.((((((..((((((((((((..((.....))..))))))))))))..))))))...((((.....))))...)))))).))...... ( -37.60) >DroSim_CAF1 81760 97 - 1 ---GAGGCUGGCGGUUGGCCGGGCGGUUUAUUAAAUCAUUUUGACAUAAUGAACCGUUAUGUCAACGGAGGGCCAGGUGCCUGAAGUCGGCGUUGGAGUC ---((.((((((.((((((..((((((((((((..((.....))..))))))))))))..))))))...((((.....))))...)))))).))...... ( -37.60) >DroEre_CAF1 89748 91 - 1 ----AAA-----GGUUGGCCGGGCGGAUUAUUAAAUCAUUUUGACAUAAUGAUCCGUUAUGUCAACGGAGGGCCAGGUGCCUGAAGUCGGCAGUGGAGUC ----...-----.((((((..((((((((((((..((.....))..))))))))))))..))))))......(((..((((.......)))).))).... ( -30.10) >DroYak_CAF1 9891 92 - 1 ---GAAA-----UGUUGCCCGCGCGGCUUAUUAAAUGAUUUUGACAUAAUGAACCGUUAUGUCAACGGAGGGCCAGGUGCUCGAAGUCGGCGUUGGAGUC ---....-----.....((.(((((((((...........(((((((((((...)))))))))))....((((.....)))).))))).)))).)).... ( -27.80) >DroAna_CAF1 10815 96 - 1 UGGUAGGUGAGAGAGUGACCGUGCUGUUUAUUAAAUCACUUUGACACAAUGAACCGUUAUGUCAACACACGGCCAGA---GCAAAGUAGUUGUUGGAGU- ((((..(((..(((((((.................)))))))((((.((((...)))).))))..)))...)))).(---((((.....))))).....- ( -19.13) >consensus ___GAGG_____GGUUGGCCGGGCGGUUUAUUAAAUCAUUUUGACAUAAUGAACCGUUAUGUCAACGGAGGGCCAGGUGCCUGAAGUCGGCGUUGGAGUC .............((((((..((((((((((((..((.....))..))))))))))))..))))))......(((.(((((.......)))))))).... (-20.48 = -22.15 + 1.67)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:38 2006