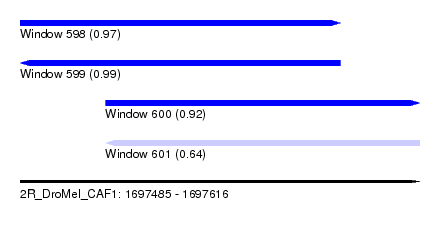
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,697,485 – 1,697,616 |
| Length | 131 |
| Max. P | 0.986936 |

| Location | 1,697,485 – 1,697,590 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1697485 105 + 20766785 AAAUGUAUAGAGG-----AUAAAAAAAUGUGUUUGUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC ............(-----(((((........))))))(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -32.60) >DroGri_CAF1 9999 103 + 1 A-A-----GAAGCAUAACAUGAA-AAAAAUUAUUGUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC .-.-----...............-.............(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -29.60) >DroSec_CAF1 83237 89 + 1 ---------------------AAAAAAAGUAUUUGUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC ---------------------................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -29.60) >DroWil_CAF1 13069 98 + 1 AAA-----CCC-------AAUAAAAAAAUCAUUUUGCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGC ...-----...-------...................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -29.60) >DroYak_CAF1 8782 96 + 1 -AAAGU------------GUUAAA-AAAGUGUUUGUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC -.....------------......-............(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -29.60) >DroMoj_CAF1 8578 96 + 1 --U-----AGCGAAC------AA-AAAAAUUAUAUCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGC --.-----.......------..-.............(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) ( -29.60) >consensus __A_______________AU_AAAAAAAGUAUUUGUCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGC .....................................(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))) (-29.60 = -29.60 + -0.00)

| Location | 1,697,485 – 1,697,590 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 82.46 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -36.90 |
| Energy contribution | -36.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1697485 105 - 20766785 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGACAAACACAUUUUUUUAU-----CCUCUAUACAUUU (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))...................-----............. ( -36.90) >DroGri_CAF1 9999 103 - 1 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGACAAUAAUUUUU-UUCAUGUUAUGCUUC-----U-U (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))((..(((((....-.....)))))...))-----.-. ( -37.30) >DroSec_CAF1 83237 89 - 1 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGACAAAUACUUUUUUU--------------------- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))................--------------------- ( -36.90) >DroWil_CAF1 13069 98 - 1 GCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGCAAAAUGAUUUUUUUAUU-------GGG-----UUU (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))((..(((((.....)))))-------..)-----).. ( -39.10) >DroYak_CAF1 8782 96 - 1 GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGACAAACACUUU-UUUAAC------------ACUUU- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))............-......------------.....- ( -36.90) >DroMoj_CAF1 8578 96 - 1 GCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGAUAUAAUUUUU-UU------GUUCGCU-----A-- (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))(((((........-.)------))))...-----.-- ( -38.80) >consensus GCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGACAAAAACUUUUUUU_AU_______________U__ (((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))..................................... (-36.90 = -36.90 + -0.00)

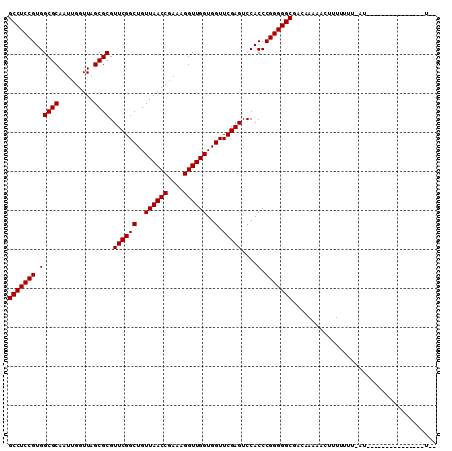
| Location | 1,697,513 – 1,697,616 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -30.62 |
| Energy contribution | -30.62 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.918438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1697513 103 + 20766785 --UGU-CGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCAUGUAAGG-------CGGAAAAAAAAUUU------UCU- --.((-((((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).......))-------)((((((....)))------)))- ( -32.11) >DroVir_CAF1 17990 103 + 1 UGAGU-CGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCCAUGUUUGC-------CGGG--UGAAUUUU------UAU- ....(-(((((((((((((.......))))).....((((((.....)))))).((((.........))))..))).(((.......))-------))))--))).....------...- ( -37.70) >DroGri_CAF1 10025 98 + 1 --UGU-CGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUGUU-------------GUA--CACCAGCUGUCUAA---- --...-.(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))).(((-------------(..--...)))).......---- ( -32.60) >DroWil_CAF1 13090 103 + 1 --UUG-CGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCGAUGUCUAC-------UCCCG-AAAAAUUU------GUAA --...-((((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).))))........-------.....-........------.... ( -33.00) >DroYak_CAF1 8801 101 + 1 --UGU-CGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCGUGGAAAC-------UUCG--AAAAAUGU------UCU- --...-.(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))((.(....)-------..))--........------...- ( -32.80) >DroAna_CAF1 9579 109 + 1 --AAUCCGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAAAAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCCACGGAGACGCUCGAGCAGCAAAAAAAUU--------CA- --..((((((....))))))(((((.((.((.....((((((.....)))))).((((.........))))...)).))...((....)).))))).............--------..- ( -36.70) >consensus __UGU_CGCCCCCGGGUGGACUCGAACCACCAACCUUUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCCAUGUAAAC_______CGGA__AAAAAUUU______UCU_ .......(((.((((((((.......))))).....((((((.....)))))).((((.........))))..))).)))........................................ (-30.62 = -30.62 + -0.00)

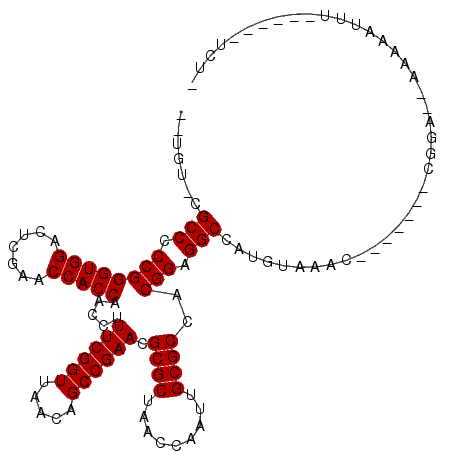
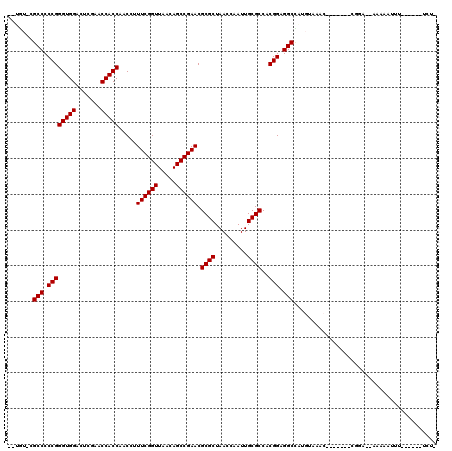
| Location | 1,697,513 – 1,697,616 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.33 |
| Mean single sequence MFE | -37.78 |
| Consensus MFE | -36.47 |
| Energy contribution | -36.47 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.639490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1697513 103 - 20766785 -AGA------AAAUUUUUUUUCCG-------CCUUACAUGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCG-ACA-- -.((------(((....)))))..-------.........(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).-...-- ( -38.50) >DroVir_CAF1 17990 103 - 1 -AUA------AAAAUUCA--CCCG-------GCAAACAUGGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCG-ACUCA -...------........--....-------.........(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).-..... ( -36.90) >DroGri_CAF1 10025 98 - 1 ----UUAGACAGCUGGUG--UAC-------------AACAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCG-ACA-- ----..............--...-------------....(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).-...-- ( -37.10) >DroWil_CAF1 13090 103 - 1 UUAC------AAAUUUUU-CGGGA-------GUAGACAUCGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCG-CAA-- ....------........-.....-------........((((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))))-...-- ( -38.10) >DroYak_CAF1 8801 101 - 1 -AGA------ACAUUUUU--CGAA-------GUUUCCACGGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCG-ACA-- -...------........--....-------.........(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).))))))).-...-- ( -36.90) >DroAna_CAF1 9579 109 - 1 -UG--------AAUUUUUUUGCUGCUCGAGCGUCUCCGUGGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUUUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCGGAUU-- -..--------..........((((((..((((..((((((((.....))).))..)))...))))..((((....(((((....)))))((((.......))))))))))))))...-- ( -39.20) >consensus _AGA______AAAUUUUU__CCCG_______GUAUACAUGGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAAAGGUUGGUGGUUCGAGUCCACCCGGGGGCG_ACA__ ........................................(((((((.(((((.........)))).((((((..((((((....))))))..).)))))....).)))))))....... (-36.47 = -36.47 + -0.00)


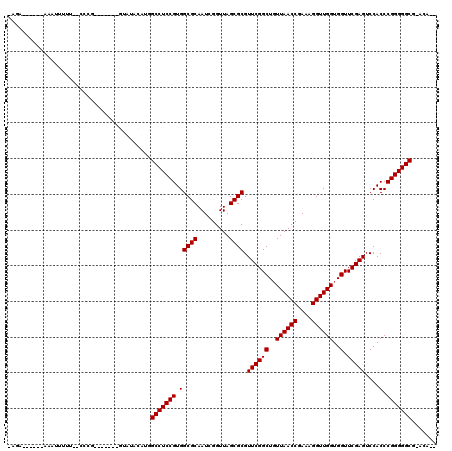
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:36 2006