| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,906,662 – 9,906,822 |
| Length | 160 |
| Max. P | 0.839299 |

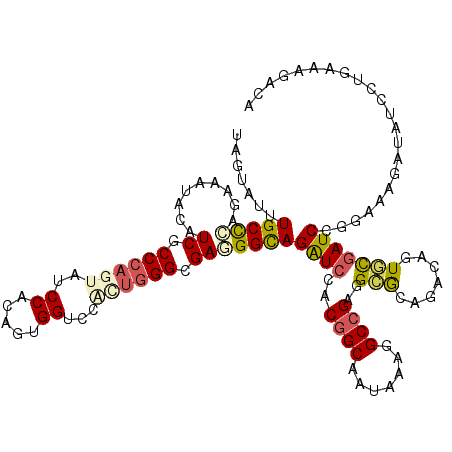
| Location | 9,906,662 – 9,906,782 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.50 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -27.66 |
| Energy contribution | -28.37 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558215 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9906662 120 + 20766785 CCUUGGGUGGGCGUAGAUUUUUCCUGGAGUCUCACGAGUAUGUCUUUCGGGAUAUCUACCAGGAUCGAAGGAUCUGCUCCUCGGCGUUCAUUGCCGUGGACCUGCCAUCGCCCAGUGGAC (((((((((((((((((((((((((((((..((.((((.......)))).))...)).))))))....)))))))))(((.(((((.....))))).)))...)))..))))))).)).. ( -51.60) >DroSec_CAF1 42791 120 + 1 CCUUGGGUGGGCGUACAUUUUUCCUGGAGUCUCACGAGUACGUCUUUCAGGAUAUCUACCAGGAUCGAAGGAUCUGCUCCUCGGCGUUCAUUGCCGUGGACUUGCCAUCGCCCAGUGGAC (((((((((((((((.((...((((((((....(((....))).)))))))).)).)))..(((((....)))))..(((.(((((.....))))).)))...)))..))))))).)).. ( -45.60) >DroEre_CAF1 33931 120 + 1 CAUUGGGUGGACGUAGAUUUUUCCUGGAGUCUCACGAGUACGUCUUUCGGGAUAUCUACCAGGAUCGAAGGAUCUGCUCCUCUGCGUUCAUUGCCGUGGACCUGCCAUCGCCCAGUGGAC ((((((((((..((((((...((((((((....(((....))).)))))))).))))))(((((((....))))...(((.(.(((.....))).).))).)))...))))))))))... ( -44.50) >DroYak_CAF1 30111 120 + 1 CGUUGGGUGGACGCACAUUCUUCCUGGAGUCUCACGAGUACGUCUUUCGGGAUAUCUACCAGGAUCGAAGGAUCUGCUCCUCCGCGUUCAUUGCCGUUGACCUGCCAUCGCCCAGUGGAC .....((.(((.(((.(((((((((((((..((.((((.......)))).))...)).))))))....))))).)))))).))..((((((((.((.((......)).))..)))))))) ( -37.30) >DroMoj_CAF1 44116 120 + 1 CCCUGGGCAAUCGUCGCUUCCAGCUGAAGGGCGAGGACUAUGUGUUCCAUGAUAUCUUUCCGGAUCGCACGGUCUGCGCCUCGGCAUUCAUUGCCGUCGAUCUGCCCUCGCCCAGCGGAC ((((((((.......((.....)).((.(((((.((((..((((.(((..((......)).))).))))..)))).((...(((((.....))))).))...))))))))))))).)).. ( -49.20) >DroAna_CAF1 29605 120 + 1 CCAUGGGAGGCCGUGCCUUCUUCCUGGAGGGCCACGAGUAUGUCUUUCGGGAUAUUUUCAAGGAUCAAAGAAUCUGCUCAUCGGCCUUUGUGGCCGUGGACCUGCCCUCCCCCCAAGGCC ((.((((((((...)))).......(((((((...(((.((((((....)))))).))).(((.(((..((......))..(((((.....)))))))).)))))))))).)))).)).. ( -45.50) >consensus CCUUGGGUGGACGUACAUUCUUCCUGGAGUCUCACGAGUACGUCUUUCGGGAUAUCUACCAGGAUCGAAGGAUCUGCUCCUCGGCGUUCAUUGCCGUGGACCUGCCAUCGCCCAGUGGAC (.((((((((..(((.(((((....))))).(((((((((.((((((((....((((....)))))))))))).)))))...((((.....))))))))...)))..)))))))).)... (-27.66 = -28.37 + 0.70)

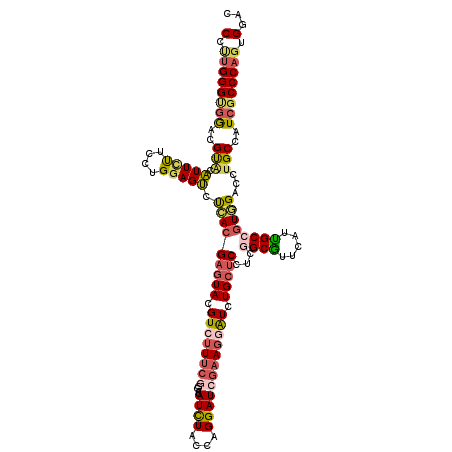
| Location | 9,906,702 – 9,906,822 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.72 |
| Mean single sequence MFE | -36.99 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.22 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.839299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9906702 120 - 20766785 UAGUAUUUGCCCAAAAACACAUCCCCCAGAAUCCAGAGCGGUCCACUGGGCGAUGGCAGGUCCACGGCAAUGAACGCCGAGGAGCAGAUCCUUCGAUCCUGGUAGAUAUCCCGAAAGACA ..(((((((((........((((.(((((...((.....))....))))).))))...(.(((.((((.......)))).))).).((((....))))..)))))))))..(....)... ( -39.20) >DroVir_CAF1 35410 120 - 1 UAGUACUUGCCCAGAAACACAUCACCCAGUAUCCACAACGGGCCACUGGGCGAGGGCAGAUCCACGGCUAUAAAUGCCGAGGCGCACACAGUGCGAUCCGAGAAAAUGUCGUGGAAUACA ..(((.((((((.(.....).((.((((((.(((.....)))..)))))).)))))))).((((((((........(.((..(((((...))))).)).).......)))))))).))). ( -43.36) >DroGri_CAF1 37303 120 - 1 UAGUAUUUGCUCAGAAAUACAUCGCCCAGUAUCCACAGUGGUCCACUUGGCGAUGGUAGAUCCACGGCUAUAAAAGCCGAUGCGCAAACUGUACGAUCCGCAAAGAUAUCAUGGAAUACA ..((((((...........(((((((.(((..((.....))...))).)))))))(..((((..(((((.....))))).((((.....)))).))))..)............)))))). ( -35.10) >DroWil_CAF1 62811 120 - 1 UAAUAUUUGCCUAGAAAUACAUCACCCAAUAUCCAUAAUGGACCACUGGGCGAAGGUAGGUCAACAGCAAUCAAGGCCGAGGCACACACAAUGCCAUCCUGAUAGGUAUCCUUAAAGAUA .......((((((((..(((.((.((((...(((.....)))....)))).))..)))..))........(((.((....((((.......))))..)))))))))))............ ( -27.40) >DroMoj_CAF1 44156 120 - 1 UAAUAUUUGCCCAGAAAUACAUCGCCCAAUAUCCAAAGUGGUCCGCUGGGCGAGGGCAGAUCGACGGCAAUGAAUGCCGAGGCGCAGACCGUGCGAUCCGGAAAGAUAUCAUGGAACACA ....(((((((((....)...(((((((....((.....)).....)))))))))))))))...(((((.....)))))..((((.....))))(.((((((......)).)))).)... ( -40.30) >DroAna_CAF1 29645 120 - 1 UAGUACUUGCCCAGGAAGACGUCGCCCAGUAUCCAUAGAGGGCCUUGGGGGGAGGGCAGGUCCACGGCCACAAAGGCCGAUGAGCAGAUUCUUUGAUCCUUGAAAAUAUCCCGAAAGACA ..(.((((((((.(.....).((.(((((..(((.....)))..))))).)).)))))))).).(((((.....))))).......(((..(((.(....).)))..))).(....)... ( -36.60) >consensus UAGUAUUUGCCCAGAAAUACAUCGCCCAGUAUCCACAGUGGUCCACUGGGCGAGGGCAGAUCCACGGCAAUAAAGGCCGAGGCGCAGACAGUGCGAUCCGGAAAGAUAUCCUGAAAGACA .......(((((.........((.((((((..((.....))...)))))).)))))))((((..((((.......))))..(((.......)))))))...................... (-20.94 = -21.22 + 0.28)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:31 2006