
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,888,839 – 9,888,985 |
| Length | 146 |
| Max. P | 0.857875 |

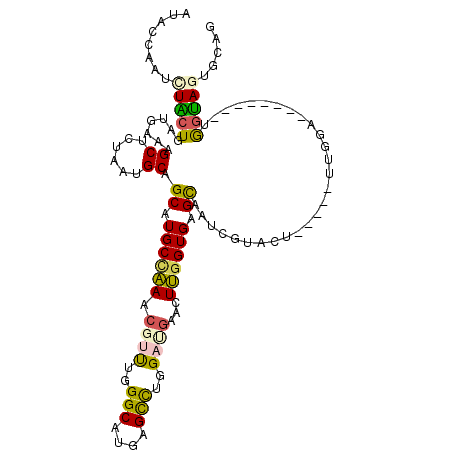
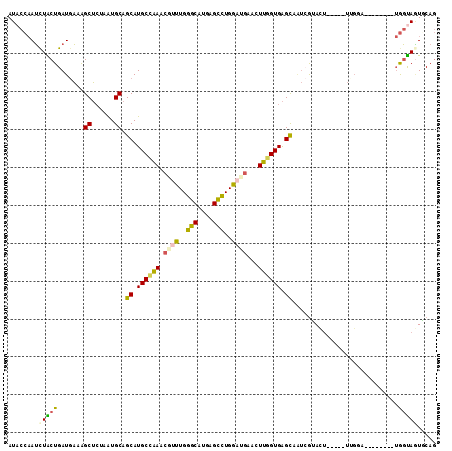
| Location | 9,888,839 – 9,888,945 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -19.60 |
| Energy contribution | -19.30 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.857875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9888839 106 - 20766785 AUACCAAUCUACUGAUGAAAGCUAUAAUGCAGCAUGCCAAACGUUUGGGCAUGAGCCUGGAUGAACUUGGUGAGCAAUCGUACU-----UUGGAUUUUGG--UGGUAAUGCAG .((((((((((..(((((..((......)).((.((((((.((((..(((....)))..))))...)))))).))..)))).).-----.)))))..)))--))......... ( -32.30) >DroSec_CAF1 12112 100 - 1 AUUCCAAUCUACUGAUGAAAGCUCUAAUGCAGCAUGCCAAUCGUUUGGGCAUGAGCCUGGAUGAACUUGGUGAGCAAUCGUAAU-----UUGGA--------UGGUAGUGCAG ..(((((..(((.(((....((......)).((.(((((((((((..(((....)))..)))))..)))))).)).))))))..-----)))))--------........... ( -32.40) >DroSim_CAF1 11232 100 - 1 AUACCAAUUUACUGAUGAAAGCUCUAGUGCAGCAUGCGAAUCGUUUGGGCAUGAGCCUGGAUGAACUUGGUGAGCAAUAGUACU-----UUGGA--------UGGUAGUGCAG ...((((..(((((.((...(((..(((((.....))...(((((..(((....)))..)))))))).)))...)).)))))..-----)))).--------........... ( -29.50) >DroEre_CAF1 16213 100 - 1 AUACCAAUCUGCUGAUGAAGGCUCUAAUGCAGCAUGCCAAACGAUUGGGCAUGAGUUUGGAGGAACUUGGUGAGCAGUCGGACU-----UCGCA--------UGGUAGUUCAG .(((((..(((((.((.(((.((((((.....((((((.........))))))...))))))...))).)).))))).((....-----.))..--------)))))...... ( -31.30) >DroYak_CAF1 12078 105 - 1 AUACCAAUCUGCUGAUGAAAGCUCUAAUGCAGCAUGCCAAUCGCUUGGGCAUGAGUUUGGAAGAACUUGGUGAGCAGUCGGACUCAAAGUAGCA--------UGGUAGUUCAG .(((((..(((((..(((..((((........((((((.........))))))(((((....)))))....))))..))).......)))))..--------)))))...... ( -28.10) >DroMoj_CAF1 14021 103 - 1 AUACACAGCUGCUGAUCUCAGCGCUCGUGCAGCAUGCUCAACGUCUGGGCAUGAGCCUGGACCAAUUGGGUGGGUAU--AAAGC-----UCUAU---UCGUAAAUCAUUUCAA ...((((((.((((....))))))).)))..((..((((((.(((..(((....)))..)))...))))))((((..--...))-----))...---..))............ ( -31.80) >consensus AUACCAAUCUACUGAUGAAAGCUCUAAUGCAGCAUGCCAAACGUUUGGGCAUGAGCCUGGAUGAACUUGGUGAGCAAUCGUACU_____UUGGA________UGGUAGUGCAG ........(((((.......((......)).((.((((((.((((..(((....)))..))))...)))))).))............................)))))..... (-19.60 = -19.30 + -0.30)

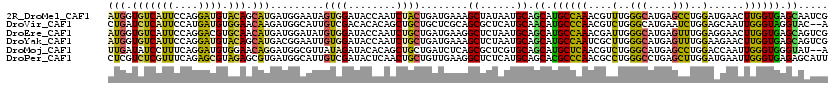
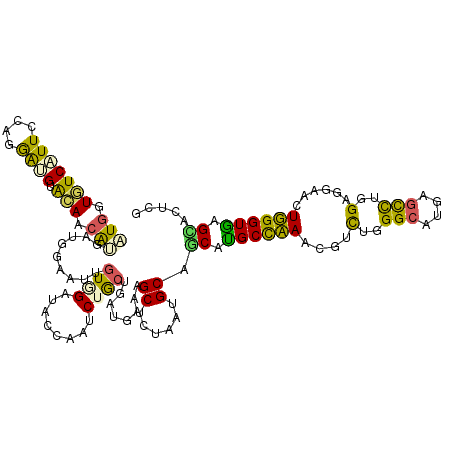
| Location | 9,888,865 – 9,888,985 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.08 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -17.83 |
| Energy contribution | -16.17 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.44 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9888865 120 - 20766785 AUGGUGUCAUUCCAGGAUGUACAGCAUGAUGGAAUAGUGGAUACCAAUCUACUGAUGAAAGCUAUAAUGCAGCAUGCCAAACGUUUGGGCAUGAGCCUGGAUGAACUUGGUGAGCAAUCG .(((((((((((((..(((.....)))..))))))....))))))).......(((....((......)).((.((((((.((((..(((....)))..))))...)))))).)).))). ( -40.10) >DroVir_CAF1 9986 118 - 1 CUGAUCUCAUUCCAUGAUGUGGAACAAGAUGGCAUUGUCGACACACAGCUGCUGCUCGCAGCGCUCAUGCAACAUGCCCAACGUCUGGGCAUGAAUCUGGAGCAAUUGGGUAGGUAC--A ...((((...((((((.((((..((((.......))))...))))))(((((.....)))))((((......((((((((.....))))))))......))))...)))).))))..--. ( -39.70) >DroEre_CAF1 16233 120 - 1 AUGGUGUCAUUCCAGGACGUGCAACAUGAUGGAUAUGUGGAUACCAAUCUGCUGAUGAAGGCUCUAAUGCAGCAUGCCAAACGAUUGGGCAUGAGUUUGGAGGAACUUGGUGAGCAGUCG .(((((((..((((...((((...)))).))))......)))))))..(((((.((.(((.((((((.....((((((.........))))))...))))))...))).)).)))))... ( -41.80) >DroYak_CAF1 12103 120 - 1 AUGGUGUCAUUCCAGGAUGUACAGCAUGACGGAAUUGUGGAUACCAAUCUGCUGAUGAAAGCUCUAAUGCAGCAUGCCAAUCGCUUGGGCAUGAGUUUGGAAGAACUUGGUGAGCAGUCG .((((((((((((...(((.....)))...)))))....)))))))..(((((.((.((..((((((.....((((((.........))))))...)))).))...)).)).)))))... ( -34.80) >DroMoj_CAF1 14046 118 - 1 UUGAUAUCCUUUCAGGAUGUGGAACAGGAUGGCGUUAUAGAUACACAGCUGCUGAUCUCAGCGCUCGUGCAGCAUGCUCAACGUCUGGGCAUGAGCCUGGACCAAUUGGGUGGGUAU--A ...((((((.(((((....(((..((((...............((((((.((((....))))))).)))...((((((((.....))))))))..))))..))).))))).))))))--. ( -42.70) >DroPer_CAF1 13872 120 - 1 CUCGUCUCGUUUCAGAGCGUAGAGCGUGAUGGCAUUGUCGAUACUCAACUGCUGUUGAAGGCUCUCAUGCAGCACGCCCAACGCCUGGGCCUGAGCUUGGAUGAAUUGGGUGAGAGCAUU ((((.((((.((((..((((.(.(((((...((((.(((.....(((((....))))).)))....))))..))))).).))))(..(((....)))..).)))).))))))))...... ( -41.50) >consensus AUGGUGUCAUUCCAGGAUGUACAACAUGAUGGAAUUGUGGAUACCAAUCUGCUGAUGAAAGCUCUAAUGCAGCAUGCCAAACGUCUGGGCAUGAGCCUGGAGGAACUGGGUGAGCACUCG (((.(((((((....)))).))).))).........((((........))))........((......)).((.((((((....(..(((....)))..)......)))))).))..... (-17.83 = -16.17 + -1.66)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:28 2006