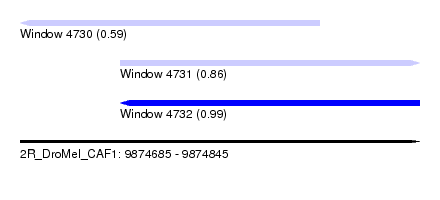
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,874,685 – 9,874,845 |
| Length | 160 |
| Max. P | 0.990026 |

| Location | 9,874,685 – 9,874,805 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.25 |
| Mean single sequence MFE | -43.72 |
| Consensus MFE | -33.88 |
| Energy contribution | -34.92 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.585152 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9874685 120 - 20766785 UCGCUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGCUCUCAUCCGCGAACACUUCUCGCCCACCAAGCCCGCCUAC ..((((((((.(....).)))).))))...(((((((..(((.(((....))).)))....((((((.(..(((....)))..).))))))..........).))))))........... ( -49.70) >DroSec_CAF1 7309 120 - 1 UCACUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGCCCUCAUCCGCGAACACUUCUCGCCCACCAAGCCCUCCUAC ....(((((..(........)..)))))..(((((((..(((.(((....))).)))....(((((((((..((....)))))..))))))..........).))))))........... ( -44.10) >DroSim_CAF1 7239 120 - 1 UCUCUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGCCCUCAUCCGCGAACACUUCUCGCCCACCAAGCCCGCCUAC ....(((((.....))))).(((..((...(((((((..(((.(((....))).)))....(((((((((..((....)))))..))))))..........).)))))).))..)))... ( -46.00) >DroYak_CAF1 7221 120 - 1 UCACUGCUGUUCUACGGAAUGGCCAGCACCUGGUGGCAGGGCGUUCGUAAGGAGGCCGUGACGUGGAAAGAUGCCAUCGAUCUCAUCCGCGACCACUUCUCGCCCACCAAGCCCGCCUAC .....(((((((....)))))))..((...(((((((..(((.(((....))).)))(((.((((((.((((.......))))..))))))..))).....).)))))).))........ ( -43.60) >DroAna_CAF1 6975 120 - 1 UCGCUCCUUUUCUACGGACUUGCCUCUAUCUGGUGGCAGGGCGUUCGCAAGGAGGCCAACACCUGGAAUGAUGCCAUUGAUCUCAUCCGGGAGCACUUCUCGCCCACAAAGCCCGCCUAC ..((((((.(((((.((..(((((.((....)).)))))(((.(((....))).)))....))))))).((((..........)))).)))))).......................... ( -35.20) >consensus UCACUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGCUCUCAUCCGCGAACACUUCUCGCCCACCAAGCCCGCCUAC ....(((((..(........)..)))))..(((((((..(((.(((....))).)))....((((((.....((....)).....))))))..........).))))))........... (-33.88 = -34.92 + 1.04)

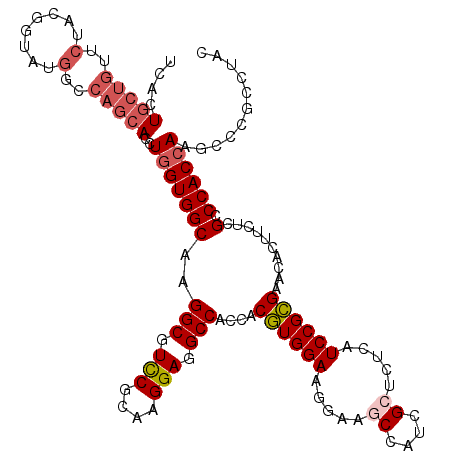
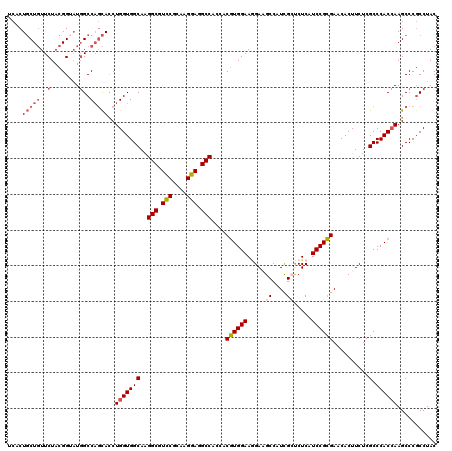
| Location | 9,874,725 – 9,874,845 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -48.04 |
| Consensus MFE | -34.44 |
| Energy contribution | -36.12 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9874725 120 + 20766785 GCGAUGGCUUCCUUCCACGUGGUGGCCUCCUUGCGGACGCCUUGCCACCAGGUGCUGGCCAUACCGUAGAACAGCAGCGAGAUGCCCUUCAGGGCGUUCUCGUCGCUGAUACCUUCUACA ...((((((......(((.(((((((((((....))).)....))))))).)))..))))))...((((((((((.(((((((((((....))))).)))))).)))).....)))))). ( -56.00) >DroSec_CAF1 7349 120 + 1 GCGAUGGCUUCCUUCCACGUGGUGGCCUCCUUGCGGACGCCUUGCCACCAGGUGCUGGCCAUACCGUAGAACAGCAGUGAGAUGCCCUUCAGGGCGUUCUCGUCGCUGAUCCCCUCCACA (((((((((......(((.(((((((((((....))).)....))))))).)))..))))))..))).((.((((..((((((((((....))))).)))))..)))).))......... ( -50.70) >DroSim_CAF1 7279 120 + 1 GCGAUGGCUUCCUUCCACGUGGUGGCCUCCUUGCGGACGCCUUGCCACCAGGUGCUGGCCAUACCGUAGAACAGCAGAGAGAUGCCCUUCAGGGCGUUCUCGUCGCUGAUCCCCUCCACA (((((((((......(((.(((((((((((....))).)....))))))).)))..))))))..))).((.((((.(((((((((((....))))).)))).)))))).))......... ( -48.60) >DroYak_CAF1 7261 120 + 1 UCGAUGGCAUCUUUCCACGUCACGGCCUCCUUACGAACGCCCUGCCACCAGGUGCUGGCCAUUCCGUAGAACAGCAGUGAGAUGCCCUUCAGGGCAUUCUCGUCGCUGAUCCCCUCCAGC ....(((.....(((.(((..(.((((((.....))..((((((....)))).)).)))).)..))).)))((((..((((((((((....))))).)))))..)))).......))).. ( -41.60) >DroAna_CAF1 7015 120 + 1 UCAAUGGCAUCAUUCCAGGUGUUGGCCUCCUUGCGAACGCCCUGCCACCAGAUAGAGGCAAGUCCGUAGAAAAGGAGCGAGAGCCCCUUGAGGGCGUUCUCGUCGGUGAUGUUCUCCAGC .....(((((((((((.((((((.((......)).)))))).((((.(......).)))).............)))(((((((((((....))).))))))))..))))))))....... ( -43.30) >consensus GCGAUGGCUUCCUUCCACGUGGUGGCCUCCUUGCGGACGCCUUGCCACCAGGUGCUGGCCAUACCGUAGAACAGCAGCGAGAUGCCCUUCAGGGCGUUCUCGUCGCUGAUCCCCUCCACA ....(((.....(((.(((..(((((((((....))).((((((....)))).)).))))))..))).)))((((.(((((((((((....))))).)))))).)))).......))).. (-34.44 = -36.12 + 1.68)

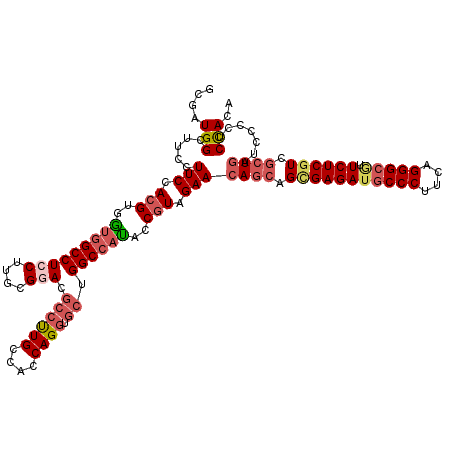
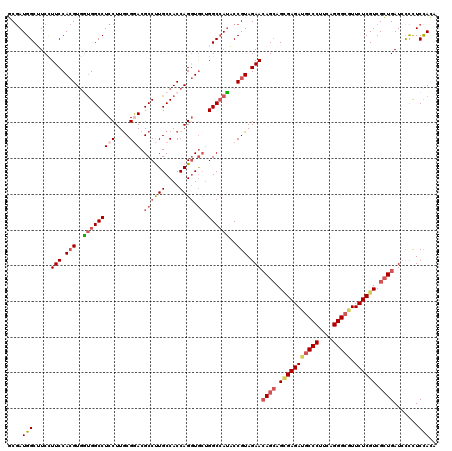
| Location | 9,874,725 – 9,874,845 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -53.24 |
| Consensus MFE | -42.18 |
| Energy contribution | -43.22 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.79 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
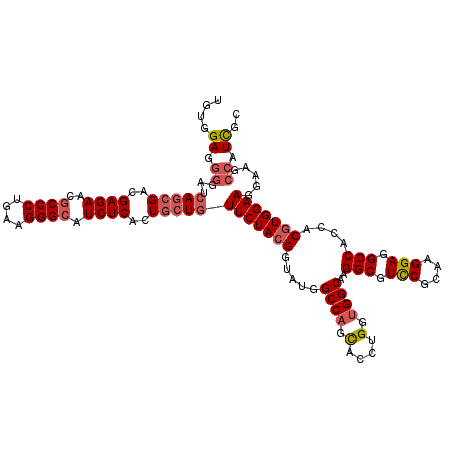
>2R_DroMel_CAF1 9874725 120 - 20766785 UGUAGAAGGUAUCAGCGACGAGAACGCCCUGAAGGGCAUCUCGCUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGC ....((.(((..(((((.(((((..((((....)))).))))).)))))(((((((.....((((.(....).))))..(((.(((....))).)))....)))))))....))).)).. ( -57.20) >DroSec_CAF1 7349 120 - 1 UGUGGAGGGGAUCAGCGACGAGAACGCCCUGAAGGGCAUCUCACUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGC .(((..((....(((((..((((..((((....)))).))))..)))))(((((((.....((((.(....).))))..(((.(((....))).)))....))))))).....))..))) ( -53.40) >DroSim_CAF1 7279 120 - 1 UGUGGAGGGGAUCAGCGACGAGAACGCCCUGAAGGGCAUCUCUCUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGC .(((..((....(((((..((((..((((....)))).))))..)))))(((((((.....((((.(....).))))..(((.(((....))).)))....))))))).....))..))) ( -53.70) >DroYak_CAF1 7261 120 - 1 GCUGGAGGGGAUCAGCGACGAGAAUGCCCUGAAGGGCAUCUCACUGCUGUUCUACGGAAUGGCCAGCACCUGGUGGCAGGGCGUUCGUAAGGAGGCCGUGACGUGGAAAGAUGCCAUCGA ....((.((.(((((((..((((.(((((....)))))))))..)))).(((((((..((((((.((.((((....)))))).(((....)))))))))..))))))).))).)).)).. ( -58.20) >DroAna_CAF1 7015 120 - 1 GCUGGAGAACAUCACCGACGAGAACGCCCUCAAGGGGCUCUCGCUCCUUUUCUACGGACUUGCCUCUAUCUGGUGGCAGGGCGUUCGCAAGGAGGCCAACACCUGGAAUGAUGCCAUUGA ..(((....(((((..(((((((..((((.....))))))))).))...(((((.((..(((((.((....)).)))))(((.(((....))).)))....))))))))))))))).... ( -43.70) >consensus UGUGGAGGGGAUCAGCGACGAGAACGCCCUGAAGGGCAUCUCACUGCUGUUCUACGGUAUGGCCAGCACCUGGUGGCAAGGCGUCCGCAAGGAGGCCACCACGUGGAAGGAAGCCAUCGC ....((.((...(((((..((((..((((....)))).))))..)))))(((((((.....((((.(....).))))..(((.(((....))).)))....))))))).....)).)).. (-42.18 = -43.22 + 1.04)

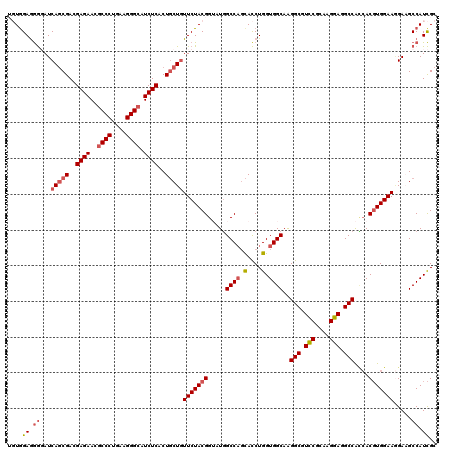
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:23 2006