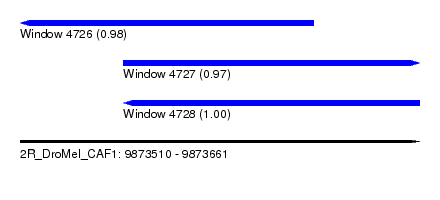
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,873,510 – 9,873,661 |
| Length | 151 |
| Max. P | 0.996760 |

| Location | 9,873,510 – 9,873,621 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.83 |
| Mean single sequence MFE | -34.11 |
| Consensus MFE | -15.71 |
| Energy contribution | -16.52 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9873510 111 - 20766785 UUGGCUAUCAAAAGAUAUGAUCACUGUGUUGAGAGACAUGGC--AAUCAUCGCUAUU------GUAACCAUAUAUUUGCUG-AUUUUUCGUCUGAUCCUGGUCCUUUGUGAAGCUAGCAA .(((.((.(((.((..(((((..(((((((....))))))).--.)))))..)).))------))).))).....((((((-.((((.((...(((....)))...)).)))).)))))) ( -26.60) >DroSec_CAF1 6134 111 - 1 CUGGCUAACAAAAGAGAUGAUCGCUCUGUUGAGAGACAUGGC--AAUCAUCGCUUUU------GUAACGAUUUAUUUGCUG-AUUUUUGGUCUGAUCCUGGUCCUCUGUGAAGCUGGCAA ...(((((((((((.((((((.(((.((((....)))).)))--.)))))).)))))------))............(((.-......((.((......)).)).......))))))).. ( -35.14) >DroYak_CAF1 5923 110 - 1 UUGGCUACCAAGAGAGAUGAUCGCUGUGUUGAGAAACAUGGC--AAUCAUCGCUUUU------GUACCAAUUU-UUUGCUC-AUCUUUCGCCUGAUCCUGGUCCUUUGUGAAGCUGGCAA ((((.(((.(((((.((((((.((((((((....))))))))--.)))))).)))))------)))))))...-.((((.(-(.(((.(((..(((....)))....)))))).)))))) ( -36.20) >DroAna_CAF1 5726 117 - 1 AUGGCAACCGAAUUUGAUGAUUUUUGCAUUCUAACAAAUGCCGAGAUCAUCCUGUUCCUUGCCGUAAUGCU---UUCGUUGAAUUUUCCGUCUGAUUCCGGUCCAUUUUGAAGGCAGCAA (((((((..((((..(((((((((.(((((......))))).)))))))))..)))).)))))))..((((---((((..((((...(((........)))...)))))))))))).... ( -38.50) >consensus UUGGCUACCAAAAGAGAUGAUCGCUGUGUUGAGAGACAUGGC__AAUCAUCGCUUUU______GUAACGAUUU_UUUGCUG_AUUUUUCGUCUGAUCCUGGUCCUUUGUGAAGCUAGCAA .........(((((.((((((.((((((((....))))))))...)))))).)))))....................((((...((((.....(((....)))......)))).)))).. (-15.71 = -16.52 + 0.81)

| Location | 9,873,549 – 9,873,661 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -33.67 |
| Consensus MFE | -15.35 |
| Energy contribution | -16.60 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.46 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9873549 112 + 20766785 AGCAAAUAUAUGGUUAC------AAUAGCGAUGAUU--GCCAUGUCUCUCAACACAGUGAUCAUAUCUUUUGAUAGCCAAAUAAUCGCUUUUUCUCCGCUGACCGGCACAAAGUGGAGCC (((...(((.(((((((------((.((..(((((.--.(..(((......)))..)..)))))..)).))).)))))).)))...)))....(((((((...........))))))).. ( -30.10) >DroSec_CAF1 6173 112 + 1 AGCAAAUAAAUCGUUAC------AAAAGCGAUGAUU--GCCAUGUCUCUCAACAGAGCGAUCAUCUCUUUUGUUAGCCAGCUAAUCGCUUUUUCUCCGCUGACCGGCACAAAGUGGAGCC (((............((------(((((.(((((((--((..(((......)))..))))))))).)))))))(((....)))...)))....(((((((...........))))))).. ( -34.80) >DroYak_CAF1 5962 111 + 1 AGCAAA-AAAUUGGUAC------AAAAGCGAUGAUU--GCCAUGUUUCUCAACACAGCGAUCAUCUCUCUUGGUAGCCAACUAAUCGCUGCUCCUCCGCCGACCGGCACAAAGUGGAGCC (((...-...(((((.(------((.((.(((((((--((..((((....))))..))))))))).)).)))...)))))......)))(((((.(.(((....))).....).))))). ( -36.80) >DroAna_CAF1 5766 117 + 1 AACGAA---AGCAUUACGGCAAGGAACAGGAUGAUCUCGGCAUUUGUUAGAAUGCAAAAAUCAUCAAAUUCGGUUGCCAUUUAAUCGUUGACCUUCCGCUGAACGGCACAAAGUGGAGGC (((((.---.(.....)(((((.(((...((((((....((((((....))))))....))))))...)))..)))))......)))))...((((((((...........)))))))). ( -33.00) >consensus AGCAAA_AAAUCGUUAC______AAAAGCGAUGAUU__GCCAUGUCUCUCAACACAGCGAUCAUCUCUUUUGGUAGCCAACUAAUCGCUGUUCCUCCGCUGACCGGCACAAAGUGGAGCC (((....................(((((.(((((((..((..(((......)))..))))))))).)))))(.....)........)))....(((((((...........))))))).. (-15.35 = -16.60 + 1.25)

| Location | 9,873,549 – 9,873,661 |
|---|---|
| Length | 112 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.96 |
| Mean single sequence MFE | -38.62 |
| Consensus MFE | -18.84 |
| Energy contribution | -19.90 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
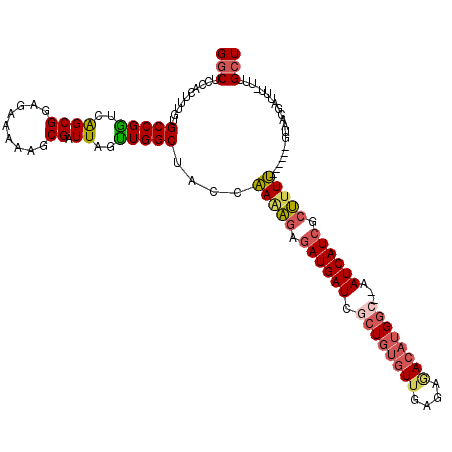
>2R_DroMel_CAF1 9873549 112 - 20766785 GGCUCCACUUUGUGCCGGUCAGCGGAGAAAAAGCGAUUAUUUGGCUAUCAAAAGAUAUGAUCACUGUGUUGAGAGACAUGGC--AAUCAUCGCUAUU------GUAACCAUAUAUUUGCU ..((((.((...........)).))))....(((((.(((.(((.((.(((.((..(((((..(((((((....))))))).--.)))))..)).))------))).))).))).))))) ( -30.60) >DroSec_CAF1 6173 112 - 1 GGCUCCACUUUGUGCCGGUCAGCGGAGAAAAAGCGAUUAGCUGGCUAACAAAAGAGAUGAUCGCUCUGUUGAGAGACAUGGC--AAUCAUCGCUUUU------GUAACGAUUUAUUUGCU (((.(......).)))(((((((................))))))).(((((((.((((((.(((.((((....)))).)))--.)))))).)))))------))............... ( -38.29) >DroYak_CAF1 5962 111 - 1 GGCUCCACUUUGUGCCGGUCGGCGGAGGAGCAGCGAUUAGUUGGCUACCAAGAGAGAUGAUCGCUGUGUUGAGAAACAUGGC--AAUCAUCGCUUUU------GUACCAAUUU-UUUGCU .(((((....(.((((....)))).))))))(((((..((((((.(((.(((((.((((((.((((((((....))))))))--.)))))).)))))------))))))))).-.))))) ( -50.00) >DroAna_CAF1 5766 117 - 1 GCCUCCACUUUGUGCCGUUCAGCGGAAGGUCAACGAUUAAAUGGCAACCGAAUUUGAUGAUUUUUGCAUUCUAACAAAUGCCGAGAUCAUCCUGUUCCUUGCCGUAAUGCU---UUCGUU ......(((((.(((......))).))))).(((((....(((((((..((((..(((((((((.(((((......))))).)))))))))..)))).)))))))......---.))))) ( -35.60) >consensus GGCUCCACUUUGUGCCGGUCAGCGGAGAAAAAGCGAUUAGUUGGCUACCAAAAGAGAUGAUCGCUGUGUUGAGAGACAUGGC__AAUCAUCGCUUUU______GUAACGAUUU_UUUGCU (((..........(((((..((((.........)).))..)))))....(((((.((((((.((((((((....))))))))...)))))).)))))....................))) (-18.84 = -19.90 + 1.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:19 2006