| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,873,016 – 9,873,128 |
| Length | 112 |
| Max. P | 0.943337 |

| Location | 9,873,016 – 9,873,128 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 62.93 |
| Mean single sequence MFE | -43.24 |
| Consensus MFE | -19.58 |
| Energy contribution | -19.86 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.803222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9873016 112 + 20766785 AUCUGCACAAGCGGCCGGUUUUGCUU---GGUGU-GGGGGCGGGGAAUGACUGUGCCGGCUGGAGG---AGCACGGCAGUGACGCGUUCCAGC-CCCUGACGUUUGUGUGUUUUGGGUUA ....((((((((((((((......))---)))..-((((((..((((((.(..((((((((.....---))).)))))..)...)))))).))-))))..)))))))))........... ( -49.10) >DroSec_CAF1 5640 112 + 1 AUCUGCACAAGCGGCCGGUUUUGCUU---GGUUU-GGGGGCGGGGAGUGACUGUGCUGGCUGGAGG---AGCACGGCAGUGACGCGUCCCAGC-CACUGGCUUUUGUGUGUUUUGGGAUG ....(((((((.(((((((.......---(((..-.((((((.(.(.((.(((((((.........---))))))))).)..).)))))).))-)))))))))))))))........... ( -43.21) >DroWil_CAF1 6599 116 + 1 CUCUGCACAAGAAGCUGGAUUUACAAUACGGUUUUGGAUAUUGGGACAGUCGCCACAAGCUGGAGAUAUGGCGUGGUGGCGACGAAUCCGAAUACCCUAAC---AGUGAGGAAUGA-AGU .(((.(((..(((((((...........)))))))((.((((.(((..((((((((..((((......))))...))))))))...))).)))).))....---.))).)))....-... ( -40.60) >DroYak_CAF1 5440 106 + 1 AUCUGCACAAGUGGCCGGUUUUGCAU---GGUG--GGGGGCGGGGAGUGUCUGUGCCGGC-----G---AGCUCGGCAGUGACGUGUCCCAAC-CCCUGGCUUCUGUGUGAUUUGUGUUA ....((((((((.(((((........---(((.--(((((..((((..(((..((((((.-----.---..).)))))..)))...))))..)-)))).))).))).)).)))))))).. ( -47.10) >DroAna_CAF1 5234 104 + 1 UUCUGCAUAG-AAGCCAGAUUUAA------GC-A-AGGGGCGGGGAGAAUCGGUGUCGGCUGGAGA---GGCUCGACCUCGAUGACGCCCAGC-CCGU---UUUUGUUGUGUUUGUGAUG ((((....))-))..(((((...(------((-(-((((((.(((...(((((.(((((((.....---))).)))).)))))....))).))-))..---.))))))..)))))..... ( -36.20) >consensus AUCUGCACAAGAGGCCGGUUUUGCAU___GGUGU_GGGGGCGGGGAGUGUCUGUGCCGGCUGGAGG___AGCACGGCAGUGACGAGUCCCAGC_CCCUGACUUUUGUGUGGUUUGGGAUA ....(((((....(((.............)))...((((((.((((..(((..((((((((........))).)))))..)))...)))).)).))))......)))))........... (-19.58 = -19.86 + 0.28)


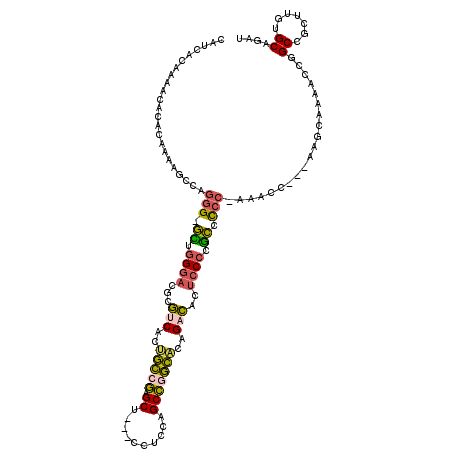
| Location | 9,873,016 – 9,873,128 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 62.93 |
| Mean single sequence MFE | -31.36 |
| Consensus MFE | -15.20 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.48 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9873016 112 - 20766785 UAACCCAAAACACACAAACGUCAGGG-GCUGGAACGCGUCACUGCCGUGCU---CCUCCAGCCGGCACAGUCAUUCCCCGCCCCC-ACACC---AAGCAAAACCGGCCGCUUGUGCAGAU ...................(((.(((-((.((((.((.....(((((.(((---.....))))))))..))..))))..))))).-.(((.---((((..........)))))))..))) ( -30.70) >DroSec_CAF1 5640 112 - 1 CAUCCCAAAACACACAAAAGCCAGUG-GCUGGGACGCGUCACUGCCGUGCU---CCUCCAGCCAGCACAGUCACUCCCCGCCCCC-AAACC---AAGCAAAACCGGCCGCUUGUGCAGAU ...................(((((((-(((((((((((....))).(((((---.........))))).))).......((....-.....---..))....)))))))).)).)).... ( -29.30) >DroWil_CAF1 6599 116 - 1 ACU-UCAUUCCUCACU---GUUAGGGUAUUCGGAUUCGUCGCCACCACGCCAUAUCUCCAGCUUGUGGCGACUGUCCCAAUAUCCAAAACCGUAUUGUAAAUCCAGCUUCUUGUGCAGAG ...-......(((((.---(((.(((((((.((((..((((((((...((..........))..)))))))).)))).)))))))..))).))............((.......)).))) ( -32.30) >DroYak_CAF1 5440 106 - 1 UAACACAAAUCACACAGAAGCCAGGG-GUUGGGACACGUCACUGCCGAGCU---C-----GCCGGCACAGACACUCCCCGCCCCC--CACC---AUGCAAAACCGGCCACUUGUGCAGAU ........(((.(((((..(((.(((-((.((((...(((..(((((.(..---.-----.))))))..)))..)))).))))).--....---..(......))))...)))))..))) ( -34.40) >DroAna_CAF1 5234 104 - 1 CAUCACAAACACAACAAAA---ACGG-GCUGGGCGUCAUCGAGGUCGAGCC---UCUCCAGCCGACACCGAUUCUCCCCGCCCCU-U-GC------UUAAAUCUGGCUU-CUAUGCAGAA ...................---..((-((.(((.(..((((..((((.((.---......))))))..)))).).))).))))..-.-..------.....((((.(..-....))))). ( -30.10) >consensus CAUCACAAAACACACAAAAGCCAGGG_GCUGGGACGCGUCACUGCCGAGCU___CCUCCAGCCGGCACAGACACUCCCCGCCCCC_AAACC___AAGCAAAACCGGCCGCUUGUGCAGAU .......................(((.((.((((...(((..(((((.((..........)))))))..)))..)))).)).)))....................((.......)).... (-15.20 = -14.80 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:14 2006