
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,843,635 – 9,843,743 |
| Length | 108 |
| Max. P | 0.815126 |

| Location | 9,843,635 – 9,843,743 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 76.82 |
| Mean single sequence MFE | -37.64 |
| Consensus MFE | -20.32 |
| Energy contribution | -20.88 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9843635 108 + 20766785 AACAGCAGCUACUACGUCCG------CACUCGCAAUCCUACCAAGCGUGUCAUGUGGAGCGCAAAUCACUGCCGAACAACUUGAGUCCCAUAAUGGUGGGUCUGCCCACUGGGA--- ..(.((((......((((((------(((.(((...........))).))...))))).)).......)))).)...........(((((.....(((((....))))))))))--- ( -29.22) >DroVir_CAF1 25769 117 + 1 AGCAACAGCUGCUGCGCUCACAAUCCCACUCGCAUUCCUAUCAGGCGCUGCACGUAGAGCGCAAAUCGCUGCCCAACAAUCUGAGUCCCAUGCUCUCGGGCAUGUCCAACAGCGGCU ......((((((((.(((((...........(((..((.....))...)))..((.(.((((.....)).)).).))....)))))..((((((....)))))).....)))))))) ( -33.00) >DroPse_CAF1 2893 95 + 1 AGCAGCAGCUGCUGCGCCCC------CACUCCCAGUCCUAUCAGGCGCUGCAUGUGGAGCGCAAGUCGCUGCCGAACAACAUGAGUCCCAUCAUGAUGGGA---------------- ....(((((.(((((((..(------(((...((((((.....)).))))...)))).)))).))).))))).............((((((....))))))---------------- ( -38.90) >DroEre_CAF1 2982 108 + 1 AACAGCAGCUGCUGCGUCCA------CACUCGCAAUCCUACCAAGCGUUGCAUGUGGAGCGCAAGUCGCUGCCGAAUAACUUGAGUCCCAUCAUGGUGGGCCUGCCCACCGGGA--- ..(.(((((.((((((((((------((...(((((.((....)).))))).)))))).))).))).))))).)...........((((.....((((((....))))))))))--- ( -46.30) >DroYak_CAF1 2881 108 + 1 AGCAGCAGCUGCUGCGUCCA------CACUCCCAAUCCUACCAAGCGUUGCAUGUGGAGCGCAAGUCGCUGCCGAAUAACUUGAGUCCUAUAAUGGUGGGCCUGCCCAUCGGCA--- .((.(((((.((((((((((------((....((((.((....)).))))..)))))).))).))).))))).....................(((((((....))))))))).--- ( -39.50) >DroPer_CAF1 6409 95 + 1 AGCAGCAGCUGCUGCGCCCC------CACUCCCAGUCCUAUCAGGCGCUGCAUGUGGAGCGCAAGUCGCUGCCGAACAACAUGAGUCCCAUCAUGAUGGGA---------------- ....(((((.(((((((..(------(((...((((((.....)).))))...)))).)))).))).))))).............((((((....))))))---------------- ( -38.90) >consensus AGCAGCAGCUGCUGCGCCCA______CACUCCCAAUCCUACCAAGCGCUGCAUGUGGAGCGCAAGUCGCUGCCGAACAACUUGAGUCCCAUCAUGGUGGGCCUGCCCA_CGG_A___ ....(((((.(((((((.(.......(((...((((.((....)).))))...)))).)))).))).)))))..............(((((....)))))................. (-20.32 = -20.88 + 0.56)

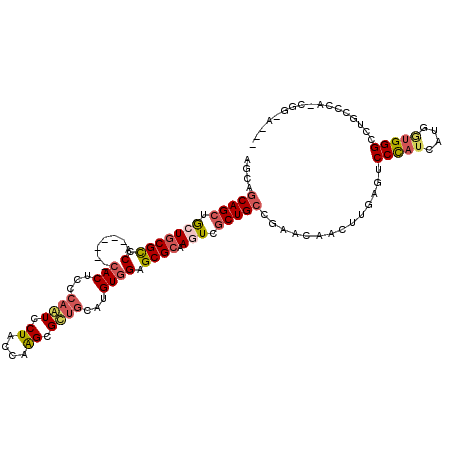
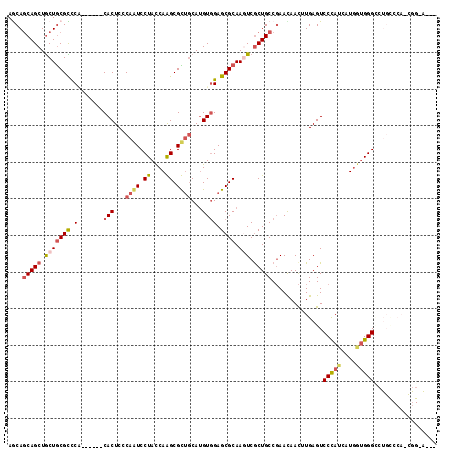
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:11 2006