| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,693,494 – 1,693,602 |
| Length | 108 |
| Max. P | 0.695102 |

| Location | 1,693,494 – 1,693,602 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 79.73 |
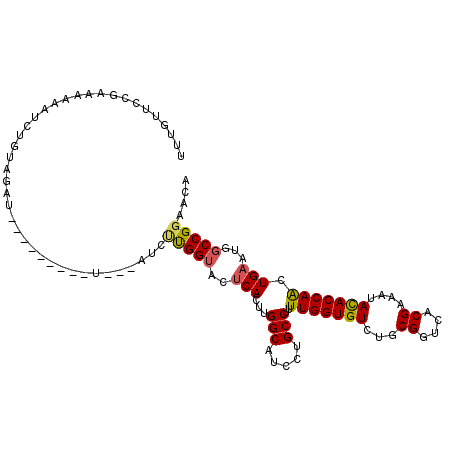
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -19.01 |
| Energy contribution | -20.40 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1693494 108 + 20766785 UUUGUCCCGAUAAAAUGUUGAGAUCUUUUUCUUU---UUUUUGGUACUCACUUGGCAUCCUGCCUUUUGGUGUCUGCGGUGACGAAAUACACCAACUGAAUGGCCGGAACA ...((.((((.((((....((((....)))).))---)).)))).))..........(((.(((..(((((((...((....))....)))))))......))).)))... ( -27.70) >DroSec_CAF1 78951 99 + 1 UUUGUUCCAAAUAAAUCUGUAGGU---------U---AUCCUGGUACUCACUUGGCAUCCUGCCUUUUGGUGUCUGCGGUGACGAAAUACACCAACUGAAUGGCCGGAACA ..((((((............(((.---------.---..)))(((..(((...(((.....)))..(((((((...((....))....))))))).)))...))))))))) ( -31.60) >DroSim_CAF1 76614 99 + 1 UUUGUUCCGAAUAAAUCUGUAGGU---------U---AUCUUGGUACUCACUUGGCAUCCUGCCUUUUGGUGUCUGCGGUGACGAAAUACACCAACUGAAUGGCCGGAACA ..((((((((.....))....(((---------(---((.(..((........(((.....)))...((((((...((....))....))))))))..))))))))))))) ( -32.00) >DroEre_CAF1 84222 98 + 1 UUGGUG-CCGAAAAAGGCGUAGGU---------C---AUUUCGGUACUCACUUGGCAUCCUGCCUCUUGGUGUCUGCGGUCACGAAAUACACCAACUGAAUGGCCGGAACA ..((((-((((((..(((....))---------)---.)))))))))).........(((.((((((((((((...((....))....)))))))..))..))).)))... ( -32.40) >DroYak_CAF1 4932 98 + 1 UGGUUU-CGAACAAAUGCCUAGAU---------A---AUUUUGGUACUCACUUGGCAUCCUGCCUUUUGGUGUCUGCGGUCACGAAAUACACCAAUUGAAUGGCCGGAACA ..((((-((..((..((((.(((.---------.---..))))))).(((...(((.....)))..(((((((...((....))....))))))).))).))..)))))). ( -26.30) >DroPer_CAF1 104703 102 + 1 UAAUCUCCGAAACAACCUAUGGAU---------UUCCGCCACGGAACUCACUUGGCAUCCUGCCGCUUGGUGUCGGCGGUCACGAAGUAUACCAGCUGUAUGGCCGGCACA ......((((....(((....((.---------(((((...))))).))....(((.....)))....))).))))((((((...(((......)))...))))))..... ( -29.20) >consensus UUUGUUCCGAAAAAAUCUGUAGAU_________U___AUCUUGGUACUCACUUGGCAUCCUGCCUUUUGGUGUCUGCGGUCACGAAAUACACCAACUGAAUGGCCGGAACA ........................................(((((..(((...(((.....)))..(((((((...((....))....))))))).)))...))))).... (-19.01 = -20.40 + 1.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:32 2006