
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,839,063 – 9,839,404 |
| Length | 341 |
| Max. P | 0.999502 |

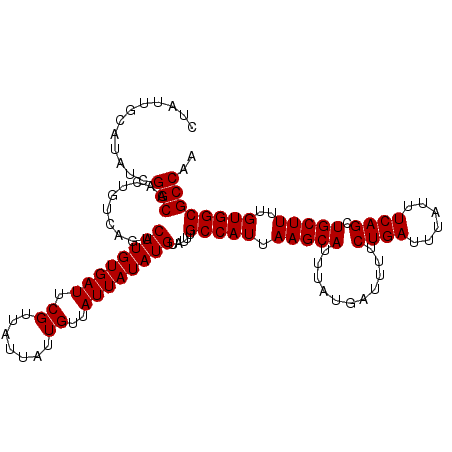
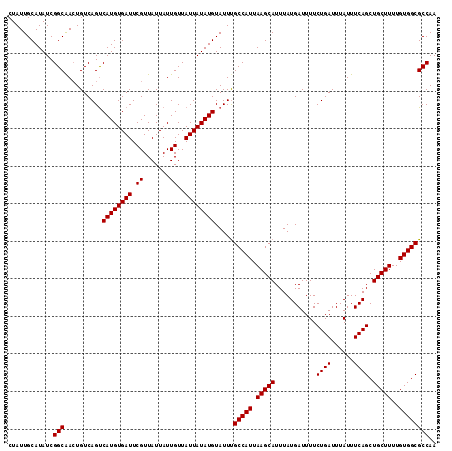
| Location | 9,839,063 – 9,839,177 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 98.60 |
| Mean single sequence MFE | -24.18 |
| Consensus MFE | -24.10 |
| Energy contribution | -24.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839063 114 + 20766785 CUAUUGCAUAUCGGCAACUGUCAGUCAUGUGAUUCGUUAUUAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAA ............(((..........((((((((.((.......))..)))))))).....(((((.(((((...........((((......)))).)))))..)))))))).. ( -24.00) >DroSec_CAF1 78175 114 + 1 CUAUUGCAUAUCGGCAACUGUCAGCCAUGUGAUUCGUUAUUAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAA ............(((..........((((((((.((.......))..)))))))).....(((((.(((((...........((((......)))).)))))..)))))))).. ( -24.20) >DroSim_CAF1 79465 114 + 1 CUAUUGCAUAUCGGCAACUGUCAGUCAUGUGAUUCGUUAUUAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAA ............(((..........((((((((.((.......))..)))))))).....(((((.(((((...........((((......)))).)))))..)))))))).. ( -24.00) >DroEre_CAF1 81242 114 + 1 CUAUUGCAUAUCGGCAACUGUCAGUCAUGUGAUUCGUUAUUAUUGUUAUUAUAUGUAUUCGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAA ............(((..........((((((((.((.......))..)))))))).....(((((.(((((...........((((......)))).)))))..)))))))).. ( -24.30) >DroYak_CAF1 78920 114 + 1 CUAUUGCAUAUCGGCGAGUGUCAGUCAUGUGAUUCGUUAUUAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAA ............((((((.(((((.((((((((.((.......))..))))))))((..(((......)))..)).......)))))....))).((..(....)..))))).. ( -24.40) >consensus CUAUUGCAUAUCGGCAACUGUCAGUCAUGUGAUUCGUUAUUAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAA ............(((..........((((((((.((.......))..)))))))).....(((((.(((((...........((((......)))).)))))..)))))))).. (-24.10 = -24.10 + -0.00)

| Location | 9,839,063 – 9,839,177 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 98.60 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -19.28 |
| Energy contribution | -19.16 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839063 114 - 20766785 UUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUAAUAACGAAUCACAUGACUGACAGUUGCCGAUAUGCAAUAG (((((((((...(((((.((((......))))...........)))))..))))......................(((...(((......)))..)))))))).......... ( -20.40) >DroSec_CAF1 78175 114 - 1 UUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUAAUAACGAAUCACAUGGCUGACAGUUGCCGAUAUGCAAUAG (((((((((...(((((.((((......))))...........)))))..))))......................(((...(((......)))..)))))))).......... ( -20.40) >DroSim_CAF1 79465 114 - 1 UUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUAAUAACGAAUCACAUGACUGACAGUUGCCGAUAUGCAAUAG (((((((((...(((((.((((......))))...........)))))..))))......................(((...(((......)))..)))))))).......... ( -20.40) >DroEre_CAF1 81242 114 - 1 UUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCGAAUACAUAUAAUAACAAUAAUAACGAAUCACAUGACUGACAGUUGCCGAUAUGCAAUAG (((((((((...(((((.((((......))))...........)))))..))))......................(((...(((......)))..)))))))).......... ( -20.40) >DroYak_CAF1 78920 114 - 1 UUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUAAUAACGAAUCACAUGACUGACACUCGCCGAUAUGCAAUAG (((((((((...(((((.((((......))))...........)))))..))))............................(((......))).....))))).......... ( -19.40) >consensus UUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUAAUAACGAAUCACAUGACUGACAGUUGCCGAUAUGCAAUAG (((((((((...(((((.((((......))))...........)))))..))))..................................(((.....)))))))).......... (-19.28 = -19.16 + -0.12)

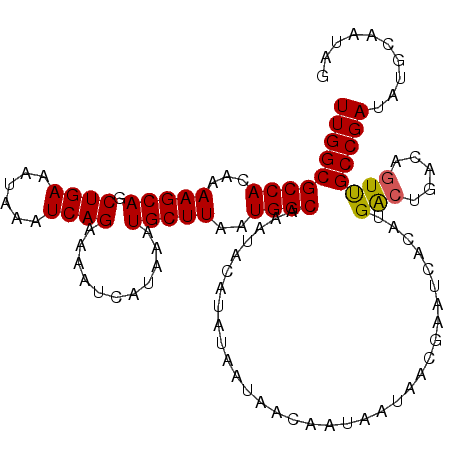
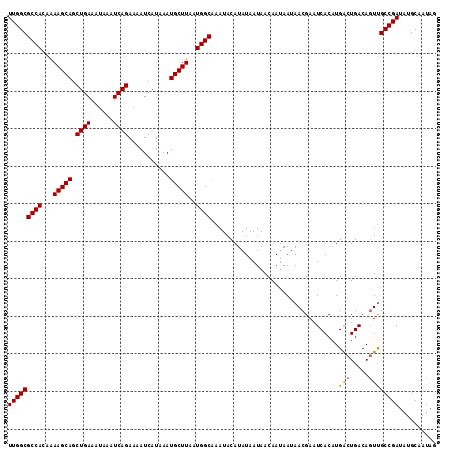
| Location | 9,839,103 – 9,839,217 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 98.04 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -22.36 |
| Energy contribution | -22.36 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.88 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839103 114 + 20766785 UAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGCAGCAGCUGCUGCCCACGUAGC ...........(((((....(((((.(((((...........((((......)))).)))))..)))))......(((.....))).....(((((....)))))..))))).. ( -26.60) >DroSec_CAF1 78215 111 + 1 UAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAGAAAGACAAUCAGC---AGCUGCUGCCCACGUAGC .((((((.......(((..(((......)))..)))...((((((((((......((..(....)..))....))))))))))))))))....---.(((((.......))))) ( -26.90) >DroSim_CAF1 79505 111 + 1 UAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAGAAAGACAAUCAGC---AGCUGCUGCCCACGUAGC .((((((.......(((..(((......)))..)))...((((((((((......((..(....)..))....))))))))))))))))....---.(((((.......))))) ( -26.90) >DroEre_CAF1 81282 111 + 1 UAUUGUUAUUAUAUGUAUUCGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGC---AGCUGCUGCCCACGUAGC ...........(((((...((((((.(((((...........((((......)))).)))))..)))))).....(((.....)))...((((---....))))...))))).. ( -23.90) >DroYak_CAF1 78960 111 + 1 UAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGC---AGCUGCUGCCCACGUAGC ...........(((((....(((((.(((((...........((((......)))).)))))..)))))......(((.....)))...((((---....))))...))))).. ( -22.70) >consensus UAUUGUUAUUAUAUGUAUUUGCCAUUAAGCAUUUAUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGC___AGCUGCUGCCCACGUAGC ...........(((((....(((((.(((((...........((((......)))).)))))..)))))((....(((.....)))...((((....))))..))..))))).. (-22.36 = -22.36 + -0.00)


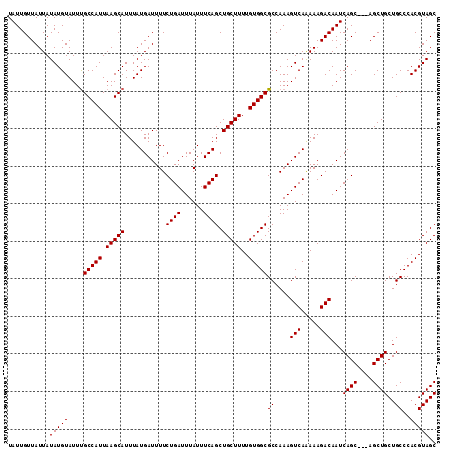
| Location | 9,839,103 – 9,839,217 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 98.04 |
| Mean single sequence MFE | -27.26 |
| Consensus MFE | -26.42 |
| Energy contribution | -26.42 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.66 |
| SVM RNA-class probability | 0.999502 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839103 114 - 20766785 GCUACGUGGGCAGCAGCUGCUGCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUA ((((.((.((((((....)))))).)).(((.....)))..))))((((...(((((.((((......))))...........)))))..)))).................... ( -29.90) >DroSec_CAF1 78215 111 - 1 GCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUCUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUA ((((..((((((.(((((---(((....(((.....)))((((......))))))))))))..........(.....).....)))))).)))).................... ( -26.60) >DroSim_CAF1 79505 111 - 1 GCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUCUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUA ((((..((((((.(((((---(((....(((.....)))((((......))))))))))))..........(.....).....)))))).)))).................... ( -26.60) >DroEre_CAF1 81282 111 - 1 GCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCGAAUACAUAUAAUAACAAUA ((((..((((((.(((((---(((....(((.....)))((((......))))))))))))..........(.....).....)))))).)))).................... ( -26.60) >DroYak_CAF1 78960 111 - 1 GCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUA ((((..((((((.(((((---(((....(((.....)))((((......))))))))))))..........(.....).....)))))).)))).................... ( -26.60) >consensus GCUACGUGGGCAGCAGCU___GCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAUAAAUGCUUAAUGGCAAAUACAUAUAAUAACAAUA ((((.((((((..((((....))))...(((.....)))....)).))))..(((((.((((......))))...........)))))..)))).................... (-26.42 = -26.42 + -0.00)

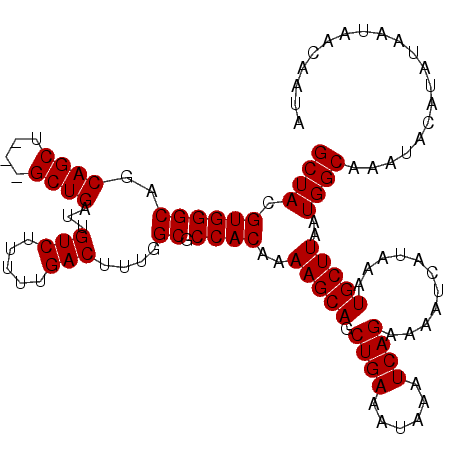
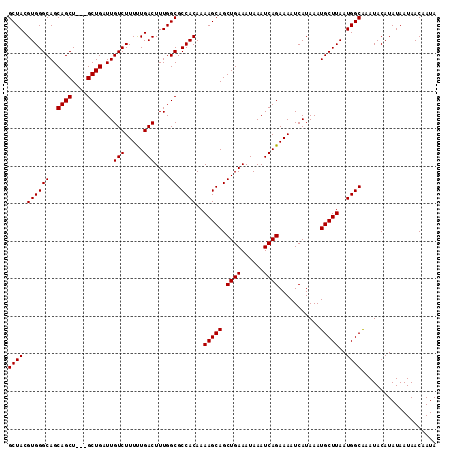
| Location | 9,839,137 – 9,839,248 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -26.46 |
| Consensus MFE | -21.98 |
| Energy contribution | -22.18 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991169 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839137 111 + 20766785 AUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGCAGCAGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAUGUGGACAUAUAUAC .(((((.((((((((......((..(....)..))....)))))....))).)))))(((((....)))))((((((((((.........))).))))))).......... ( -29.00) >DroSec_CAF1 78249 94 + 1 AUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAGAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAU-------------- .((.(((((((((((......((..(....)..))....))))))))))).))......---((((((.......))))))................-------------- ( -24.10) >DroSim_CAF1 79539 94 + 1 AUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAGAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAU-------------- .((.(((((((((((......((..(....)..))....))))))))))).))......---((((((.......))))))................-------------- ( -24.10) >DroEre_CAF1 81316 102 + 1 AUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUACCAACAAAUGUAGAAAC--ACAUAU---- (((..((((((((((.....((((((((((.((((......))))...))).....)))---))))(((((....))))).......)))).)))))).--.)))..---- ( -23.70) >DroYak_CAF1 78994 106 + 1 AUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUGCCAACAAAAGUAGAUAU--ACAUAUGUAC (((.....((((......))))(((((((((((((((....(((.....))).....))---((((((.......))))))))).))))))))))....--.)))...... ( -31.40) >consensus AUGAUUUUCUGAUUUAUUUCAGCUGCUUUUGUGGCGCCAAAGUCAAAAAGACAAUCAGC___AGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAU____ACAUAU____ ........((((......)))).(((((((((((.......(((.....)))..........((((((.......)))))).)).)))))))))................. (-21.98 = -22.18 + 0.20)


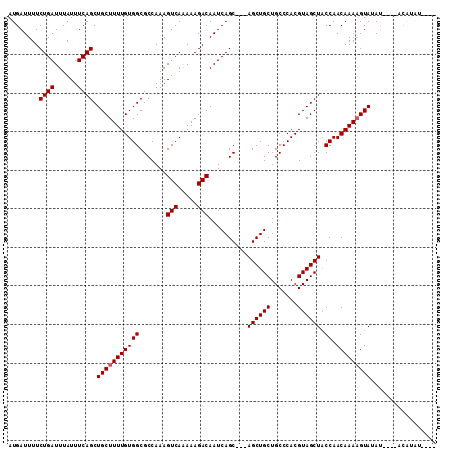
| Location | 9,839,137 – 9,839,248 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.58 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -25.68 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839137 111 - 20766785 GUAUAUAUGUCCACAUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCUGCUGCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAU ...((((((....))))))(((((((.(((.((((.((.((((((....)))))).)).(((.....)))..))))))))))))))...((((......))))........ ( -33.30) >DroSec_CAF1 78249 94 - 1 --------------AUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUCUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAU --------------.....(((((((.(((.((((.((.(((((...))---))).)).(((.....)))..))))))))))))))...((((......))))........ ( -24.50) >DroSim_CAF1 79539 94 - 1 --------------AUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUCUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAU --------------.....(((((((.(((.((((.((.(((((...))---))).)).(((.....)))..))))))))))))))...((((......))))........ ( -24.50) >DroEre_CAF1 81316 102 - 1 ----AUAUGU--GUUUCUACAUUUGUUGGUAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAU ----..(((.--.(((((..(((((((....(((.....)))..(((((---(((....(((.....)))((((......)))))))))))).))))))).)))))..))) ( -26.30) >DroYak_CAF1 78994 106 - 1 GUACAUAUGU--AUAUCUACUUUUGUUGGCAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAU ((((....))--))..((.(((((((.(((.((((.((.(((((...))---))).)).(((.....)))..)))))))))))))).))((((......))))........ ( -28.30) >consensus ____AUAUGU____AUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCU___GCUGAUUGUCUUUUUGACUUUGGCGCCACAAAAGCAGCUGAAAUAAAUCAGAAAAUCAU ...................(((((((.(((.((((((.((((((((((....)))).))))))...))....))))))))))))))...((((......))))........ (-25.68 = -25.72 + 0.04)


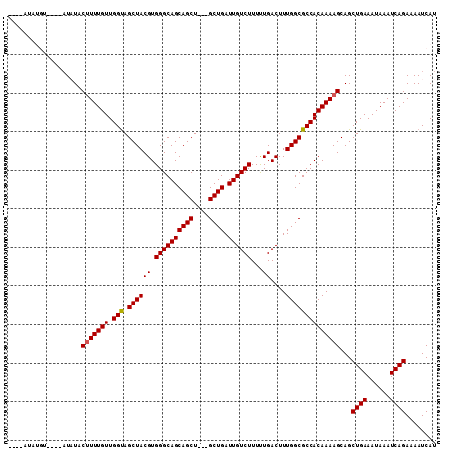
| Location | 9,839,177 – 9,839,270 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.24 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -10.50 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.58 |
| SVM decision value | 3.45 |
| SVM RNA-class probability | 0.999227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839177 93 + 20766785 AGUCAAAAAGACAAUCAGCAGCAGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAUGUGGACAUAUAUAC------------------AUAUACAUAAAAAUAUCGUUGA .(((.....)))..(((((...((((((.......))))))..........(((((((((........)))------------------))))))...........))))) ( -23.80) >DroSec_CAF1 78289 76 + 1 AGUCAGAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAU--------------------------------AUAUACAUAAAUAUAUCGUUGA .(((.....)))..(((((---((((((.......))))))..........((((..--------------------------------..))))...........))))) ( -15.70) >DroSim_CAF1 79579 76 + 1 AGUCAGAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAU--------------------------------AUAUACAUAAAUAUAUCGUUGA .(((.....)))..(((((---((((((.......))))))..........((((..--------------------------------..))))...........))))) ( -15.70) >DroEre_CAF1 81356 88 + 1 AGUCAAAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUACCAACAAAUGUAGAAAC--ACAUAU------------------ACAUAAAUACAUAUAAAUAUCUUUGA .(((.....)))..((((.---((((((.......))))))........(((((.....--.....)------------------))))..................)))) ( -11.80) >DroYak_CAF1 79034 106 + 1 AGUCAAAAAGACAAUCAGC---AGCUGCUGCCCACGUAGCUGCCAACAAAAGUAGAUAU--ACAUAUGUACAUAUACUCGCAUAGACAUAAAAAUAUAAAAAUAUCGUUGA .(((.....))).....((---((((((.......))))))))((((....(((....)--)).(((((...(((......))).)))))................)))). ( -23.10) >consensus AGUCAAAAAGACAAUCAGC___AGCUGCUGCCCACGUAGCUACCAACAAAAGUAUAU____ACAUAU______________________AUAUACAUAAAAAUAUCGUUGA .(((.....)))..(((((...((((((.......))))))....((....)).....................................................))))) (-10.50 = -10.70 + 0.20)

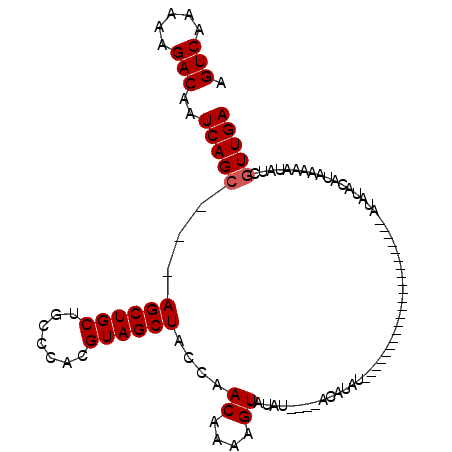
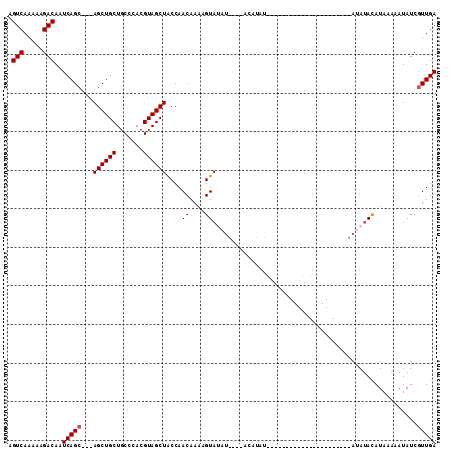
| Location | 9,839,177 – 9,839,270 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.24 |
| Mean single sequence MFE | -19.78 |
| Consensus MFE | -11.96 |
| Energy contribution | -12.16 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839177 93 - 20766785 UCAACGAUAUUUUUAUGUAUAU------------------GUAUAUAUGUCCACAUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCUGCUGCUGAUUGUCUUUUUGACU (((((((.........((((((------------------((..........))))))))..))))))).......((.((((((....)))))).)).(((.....))). ( -22.90) >DroSec_CAF1 78289 76 - 1 UCAACGAUAUAUUUAUGUAUAU--------------------------------AUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUCUGACU ......((((((....))))))--------------------------------..........((..((((((.(.......).))))---))..)).(((.....))). ( -15.20) >DroSim_CAF1 79579 76 - 1 UCAACGAUAUAUUUAUGUAUAU--------------------------------AUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUCUGACU ......((((((....))))))--------------------------------..........((..((((((.(.......).))))---))..)).(((.....))). ( -15.20) >DroEre_CAF1 81356 88 - 1 UCAAAGAUAUUUAUAUGUAUUUAUGU------------------AUAUGU--GUUUCUACAUUUGUUGGUAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUUUGACU ..(((((((..(((((((((....))------------------))))))--)...........((..((((((.(.......).))))---))..)))))))))...... ( -20.40) >DroYak_CAF1 79034 106 - 1 UCAACGAUAUUUUUAUAUUUUUAUGUCUAUGCGAGUAUAUGUACAUAUGU--AUAUCUACUUUUGUUGGCAGCUACGUGGGCAGCAGCU---GCUGAUUGUCUUUUUGACU .(((((((((............))))).....(((((.((((((....))--)))).)))))..))))((((((.(.......).))))---)).....(((.....))). ( -25.20) >consensus UCAACGAUAUUUUUAUGUAUAU______________________AUAUGU____AUAUACUUUUGUUGGUAGCUACGUGGGCAGCAGCU___GCUGAUUGUCUUUUUGACU (((((((.......................................................)))))))......((.((((((((((....)))).))))))...))... (-11.96 = -12.16 + 0.20)

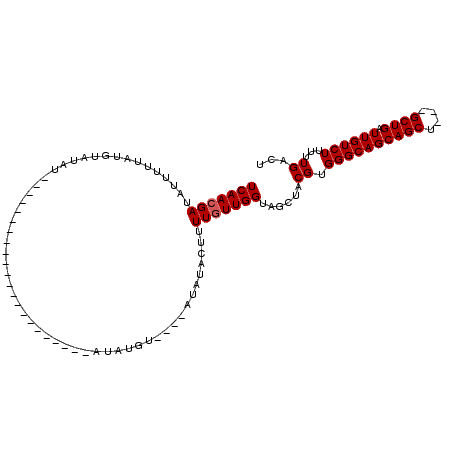

| Location | 9,839,248 – 9,839,348 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -19.73 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.933411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839248 100 + 20766785 ------------------AUAUACAUAAAAAUAUCGUUGAAAAAACCCUCGACAAUACCCAAUCUCAAUGCAAUCGAGACUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCU ------------------.................(((((........))))).........((((.........))))..(((((((((((((......))))))..)))))))... ( -20.70) >DroSec_CAF1 78343 100 + 1 ------------------AUAUACAUAAAUAUAUCGUUGAAAAAACCCUCGACAAUACCCAAUCUCAAUGCAAUCGAGACUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCU ------------------.................(((((........))))).........((((.........))))..(((((((((((((......))))))..)))))))... ( -20.70) >DroSim_CAF1 79633 100 + 1 ------------------AUAUACAUAAAUAUAUCGUUGAAAAAACCCUCGACAAUACCCAAUCUCAAUGCAAUCGAGACUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCU ------------------.................(((((........))))).........((((.........))))..(((((((((((((......))))))..)))))))... ( -20.70) >DroEre_CAF1 81418 104 + 1 --------------ACAUAAAUACAUAUAAAUAUCUUUGAAAAAACCCUCCGCAAUACCCAAUCUCAAUGCAAUCGAGACUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCU --------------.....................................(((..............))).....((((..((((((((((((......))))))..)))))))))) ( -17.14) >DroYak_CAF1 79100 118 + 1 AUAUACUCGCAUAGACAUAAAAAUAUAAAAAUAUCGUUGAAAAAACCCUCCACAAUACCCAAUCGCAAUGCAAUCGAGACUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCU ........((((.(.(......((((....)))).((.((........)).))...........)).)))).....((((..((((((((((((......))))))..)))))))))) ( -19.40) >consensus __________________AUAUACAUAAAAAUAUCGUUGAAAAAACCCUCGACAAUACCCAAUCUCAAUGCAAUCGAGACUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCU ...................................(((((........))))).......................((((..((((((((((((......))))))..)))))))))) (-17.94 = -18.38 + 0.44)

| Location | 9,839,309 – 9,839,404 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 95.48 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -20.30 |
| Energy contribution | -20.70 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972223 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9839309 95 + 20766785 CUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCUUGUAACAUU------UCGUUUUCGUUAUUUUGAACGUUAGUUUUAUUUUGAGUCGACUUAUA ...(((((((((((((((....))))...))))))).)))).......------(((..((((..((...((((....)))).))..)))).)))...... ( -22.40) >DroSec_CAF1 78404 95 + 1 CUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCUUGUAACAUU------UCGUUUGCGUUAUUUUGAACGUUAGUUUUAUUUUGAGUCGACUUAUA ...(((((((((((((((....))))...))))))).)))).......------..(((.((........((((....))))........)).)))..... ( -21.29) >DroSim_CAF1 79694 95 + 1 CUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCUUGUAACAUU------UCGUUUGCGUUAUUUUGAACGUUAGUUUUAUUUUGAGUCGACUUAUA ...(((((((((((((((....))))...))))))).)))).......------..(((.((........((((....))))........)).)))..... ( -21.29) >DroEre_CAF1 81483 101 + 1 CUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCUUGUAACGUUUCUUUUUCGUUUUCGUUAUAUUGAACGUUAGUUUUAUUUUGAGUCGACUUAUA ...(((((((((((((((....))))...))))))).))))((((((((......((....)).......)))))))).........((((....)))).. ( -24.12) >DroYak_CAF1 79179 95 + 1 CUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCUUGUAACGUU------UCGUUUUCGUUAUUUUGAACGUUAGUUUUAUUUUGAGCCGACUUAUA ..(((((((((((((......))))))..)))))))((..((((((..------.......))))))...))..((..((((......))))..))..... ( -23.20) >consensus CUUCAAGCCAAGUCAGUCAAGUGGCUUAAGGCUUGGUCUUGUAACAUU______UCGUUUUCGUUAUUUUGAACGUUAGUUUUAUUUUGAGUCGACUUAUA ...(((((((((((((((....))))...))))))).)))).............(((..((((..((...((((....)))).))..)))).)))...... (-20.30 = -20.70 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:41:06 2006