
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,837,581 – 9,837,690 |
| Length | 109 |
| Max. P | 0.841239 |

| Location | 9,837,581 – 9,837,690 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -15.77 |
| Consensus MFE | -14.10 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.64 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.609216 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
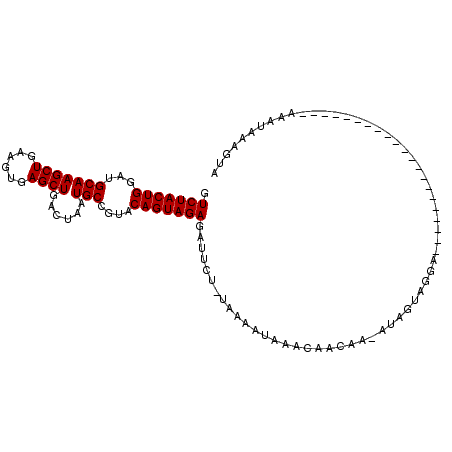
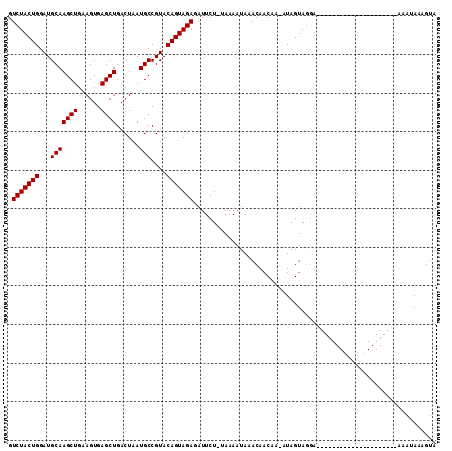
>2R_DroMel_CAF1 9837581 109 + 20766785 GUCUACUGGAUGCAAGCUGAAGUGAGCUGACUAAUGCCGUACAGUAGAGAUUCUGUAAAAUAAACAUCGA--UAGUAUAAAGUAUACAGUAAAGUAUAUAGCAAUAAAGUA .(((((((...(((((((......))))......)))....)))))))......................--.........((((((......))))))............ ( -18.00) >DroSec_CAF1 76702 89 + 1 GUCUACUGGAUGCAAGCUGAAGUGAGCUGACUAAUGCCGUACAGUAGAGAUUCU-GAAAAUAAACAACAAUAUAGUAGGA---------------------AAAUAAAGUA .(((((((...(((((((......))))......)))....)))))))..((((-(...(((........)))..)))))---------------------.......... ( -15.10) >DroSim_CAF1 78030 90 + 1 GUCUACUGGAUGCAAGCUGAAGUGAGCUGACUAAUGCCGUACAGUAGAGAUUCUUUAAAAUAAACAACAAAAUAGUAGGA---------------------AAAUAAAGUA .(((((((...(((((((......))))......)))....)))))))..(((((.....................))))---------------------)......... ( -14.20) >consensus GUCUACUGGAUGCAAGCUGAAGUGAGCUGACUAAUGCCGUACAGUAGAGAUUCU_UAAAAUAAACAACAA_AUAGUAGGA_____________________AAAUAAAGUA .(((((((...(((((((......))))......)))....)))))))............................................................... (-14.10 = -14.10 + -0.00)

| Location | 9,837,581 – 9,837,690 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.13 |
| Mean single sequence MFE | -15.66 |
| Consensus MFE | -13.10 |
| Energy contribution | -13.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841239 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
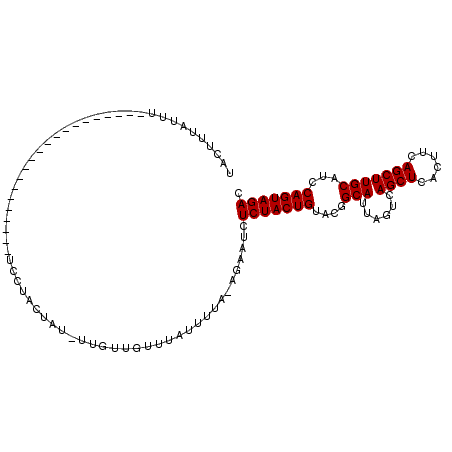
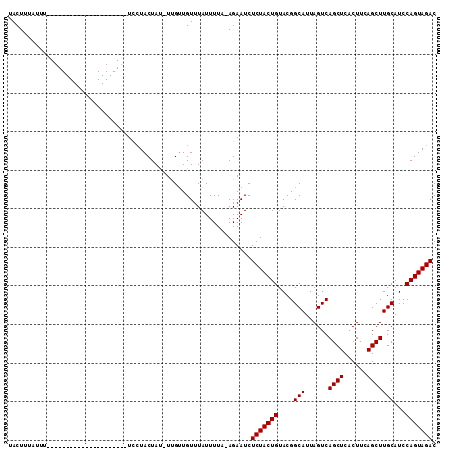
>2R_DroMel_CAF1 9837581 109 - 20766785 UACUUUAUUGCUAUAUACUUUACUGUAUACUUUAUACUA--UCGAUGUUUAUUUUACAGAAUCUCUACUGUACGGCAUUAGUCAGCUCACUUCAGCUUGCAUCCAGUAGAC ...(((((((...(((((......)))))..........--..(((((......(((((........))))).(((....)))((((......)))).)))))))))))). ( -17.80) >DroSec_CAF1 76702 89 - 1 UACUUUAUUU---------------------UCCUACUAUAUUGUUGUUUAUUUUC-AGAAUCUCUACUGUACGGCAUUAGUCAGCUCACUUCAGCUUGCAUCCAGUAGAC ..........---------------------..(((((.((.((((((.......(-((........))).)))))).))((.((((......)))).))....))))).. ( -14.50) >DroSim_CAF1 78030 90 - 1 UACUUUAUUU---------------------UCCUACUAUUUUGUUGUUUAUUUUAAAGAAUCUCUACUGUACGGCAUUAGUCAGCUCACUUCAGCUUGCAUCCAGUAGAC ..(((((...---------------------....((......)).........)))))....(((((((....(((......((((......)))))))...))))))). ( -14.69) >consensus UACUUUAUUU_____________________UCCUACUAU_UUGUUGUUUAUUUUA_AGAAUCUCUACUGUACGGCAUUAGUCAGCUCACUUCAGCUUGCAUCCAGUAGAC ...............................................................(((((((....(((......((((......)))))))...))))))). (-13.10 = -13.10 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:54 2006