
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,835,008 – 9,835,127 |
| Length | 119 |
| Max. P | 0.500000 |

| Location | 9,835,008 – 9,835,127 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.22 |
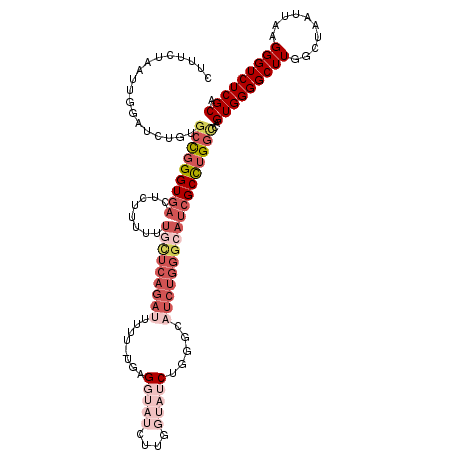
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -25.56 |
| Energy contribution | -29.00 |
| Covariance contribution | 3.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
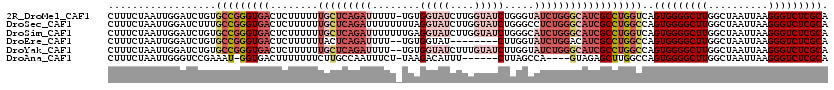
>2R_DroMel_CAF1 9835008 119 + 20766785 CUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUUU-UGUGGUAUCUUGGUAUCUGGGUAUCUGGGCAUCGCCUGGUCAGUGGGGCUUGGCUAAUUAAGGGUCUCGCA ...............((.(((((((((........(((((((((.(((-...(((((....))))).))).))))))))))))))))))))(((((((((..........))))))))). ( -39.50) >DroSec_CAF1 72409 120 + 1 CUUUCUAAUUGGAUCUUUGCCGGGUGACUCUUUUUUGCUCAGAUUUUUUUUAGGUAUCUUGGUAUCUGGGCCUCUGGGCAUCGCCUGGCCAGUGGGGCUUGGCUAAUUAAGGGUCUCGCA ..................(((((((((........((((((((.....(((((((((....)))))))))..)))))))))))))))))..(((((((((..........))))))))). ( -41.60) >DroSim_CAF1 74259 120 + 1 CUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUUUUUGAGGUAUCUUGGUAUCUGGGCAUCUGGGCAUCGCCUGGUCAGUGGGGCUUGGCUAAUUAAGGGUCUCGCA ...............((.(((((((((..(((...(((((((((....(..((....))..).)))))))))...)))..)))))))))))(((((((((..........))))))))). ( -42.40) >DroEre_CAF1 77391 110 + 1 CUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUACUCAGAUUUU--UGUGGUAU--------CUUGGUAUCUGGACAUCGCCUGGCCAGUGGGGCUUGGCUAAUUAAGGGUCUCGCA ...............((.(((((((((.(((....((((.((((...--......))--------)).))))...)))..)))))))))))(((((((((..........))))))))). ( -34.10) >DroYak_CAF1 74880 118 + 1 CUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUU--UGUGGUAUCUUUGUAUCUUGGUAUCUGGGCAUCGCCUGGCCAGUGGGGCUUGGCUAAUUAAGGGUCUCGCA ...............((.(((((((((........(((((((((...--...(((((....))))).....))))))))))))))))))))(((((((((..........))))))))). ( -40.80) >DroAna_CAF1 72212 108 + 1 CUUUCUAAUUGGGUCCGAAAU-GGUGACUUUUUUUCUUGCCAAUUUCU-UAAGACAUUU------CUUAGCCA----GUAGAGCUUGGCCAGUGGGGCUUGGCUAAUUAAGGGUCUCGCA ..........((..(((((((-((..(.........)..)).))))).-..........------.(((((((----....(((((.(....).))))))))))))....))..)).... ( -25.80) >consensus CUUUCUAAUUGGAUCUGUGCCGGGUGACUCUUUUUUGCUCAGAUUUUU_UGAGGUAUCUUGGUAUCUGGGCAUCUGGGCAUCGCCUGGCCAGUGGGGCUUGGCUAAUUAAGGGUCUCGCA ..................(((((((((........(((((((((........(((((....))))).....))))))))))))))))))..(((((((((..........))))))))). (-25.56 = -29.00 + 3.45)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:48 2006