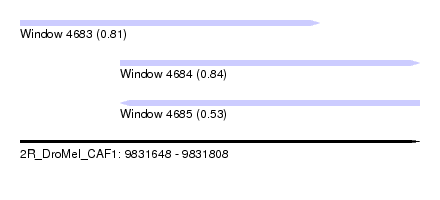
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,831,648 – 9,831,808 |
| Length | 160 |
| Max. P | 0.841127 |

| Location | 9,831,648 – 9,831,768 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -36.63 |
| Consensus MFE | -28.42 |
| Energy contribution | -30.43 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9831648 120 + 20766785 GGCCGCUGUGUAAUCCCAACAUUGGAAGCCAUCCGAUGCUCCACUGCUCCACUGUGUGUCAGUGAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCUUCUAAAAUGCAAAAUAAUUUACC ....(((((.........)))((((((((.((((((...((((.(..((((.((((((.((.....)).)))))).)))).).)))))))))).))))))))...))............. ( -35.80) >DroSec_CAF1 69143 101 + 1 GGCCGCUGUGUAAUCCCACCGUUGGAAGCCAUCCGAUGCUC--------CACUGUGUGUCAGUGAAUGCCACACGUU-----------UGGUUUGCUUCUAAAAUGCAAAAUAAUUUACC .((((.(((((........(((((((.....)))))))...--------(((((.....)))))......)))))..-----------))))((((.........))))........... ( -22.20) >DroEre_CAF1 74115 120 + 1 GGCCGCUGUGUAAUCCCGCCAUCGGCCGCCAUUCGAUGCUCCACUGCUCCACUGUGUGGCAGUAAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCGUCUGAAAUGCAAAAUAAUUUACC (((((.((.((......)))).)))))((..((((((((((((.(..((((.(((((((((.....))))))))).)))).).)))).......))))).)))..))............. ( -44.51) >DroYak_CAF1 71486 120 + 1 GGCCGCUGUGUAAUCCCCCCAUUGGCCGCCAAUCGAUGCUCCACUGCUCCACUGUGUGGCAGUGAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCGUCUAAAAUGCAAAAUAAUUUACC (((((.((.(........))).))))).((((((.((((......))((((.(((((((((.....))))))))).))))..)).)))))).((((((.....))))))........... ( -44.00) >consensus GGCCGCUGUGUAAUCCCACCAUUGGAAGCCAUCCGAUGCUCCACUGCUCCACUGUGUGGCAGUGAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCGUCUAAAAUGCAAAAUAAUUUACC ....((.(((......)))..((((((((.((((((...((((....((((.(((((((((.....))))))))).))))...)))))))))).))))))))...))............. (-28.42 = -30.43 + 2.00)



| Location | 9,831,688 – 9,831,808 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -30.54 |
| Consensus MFE | -21.04 |
| Energy contribution | -24.94 |
| Covariance contribution | 3.90 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.841127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9831688 120 + 20766785 CCACUGCUCCACUGUGUGUCAGUGAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCUUCUAAAAUGCAAAAUAAUUUACCAAAUUAACUGUACCAUUAUCAACAGCGACGAUCCGAUCAA .......((((.((((((.((.....)).)))))).)))).....(((((((((((...............(((((.....))))).((((..........)))).).)))))))))).. ( -27.80) >DroSec_CAF1 69183 101 + 1 C--------CACUGUGUGUCAGUGAAUGCCACACGUU-----------UGGUUUGCUUCUAAAAUGCAAAAUAAUUUACCAAAUUAACGGUACCAUUAUCAACAGCGACGAUCCGAUCAA .--------(.((((.((..((((..((((....(((-----------((((((((.........))))........)))))))....)))).))))..)))))).)............. ( -18.20) >DroEre_CAF1 74155 120 + 1 CCACUGCUCCACUGUGUGGCAGUAAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCGUCUGAAAUGCAAAAUAAUUUACCAAAUUAACGGUACCAUUAUCAACAGCGACGAUCCGAUCAA .......((((.(((((((((.....))))))))).)))).....((((((((((((.(((((((........))))(((........)))...........))))).)))))))))).. ( -41.50) >DroYak_CAF1 71526 120 + 1 CCACUGCUCCACUGUGUGGCAGUGAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCGUCUAAAAUGCAAAAUAAUUUACCAAAUUAACGGUACCAUUAUCAACAGCGACGAUCCGAUCAA .......((((.(((((((((.....))))))))).)))).....(((((((((((((......((....(((((.((((........))))..)))))...))))).)))))))))).. ( -38.80) >DroAna_CAF1 68871 109 + 1 -----------GGGCAUGGCCGUGAAUGCCACACAUUGGAAAAUGGAUUGGUUUGCGUCUAAAAUGCAAAAUAAUUUACCAAAUUAACGGUACCAUUAUCAACAGCGACGAUCCGAUCAA -----------.(((((........)))))...((((....))))((((((....((((.....((....(((((.((((........))))..)))))...))..))))..)))))).. ( -26.40) >consensus CCACUGCUCCACUGUGUGGCAGUGAAUGCCACACGUUGGAAAAUGGAUUGGGUUGCGUCUAAAAUGCAAAAUAAUUUACCAAAUUAACGGUACCAUUAUCAACAGCGACGAUCCGAUCAA .......((((.(((((((((.....))))))))).)))).....(((((((((((((......((....(((((.((((........))))..)))))...))))).)))))))))).. (-21.04 = -24.94 + 3.90)

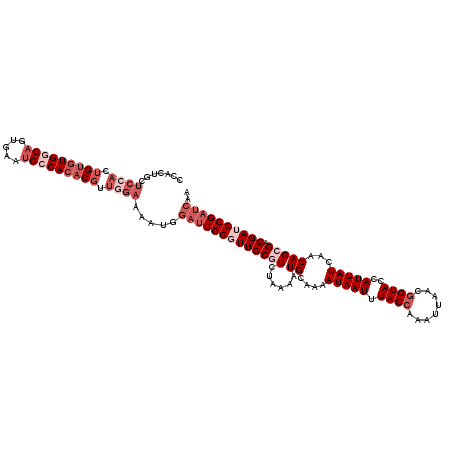
| Location | 9,831,688 – 9,831,808 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.74 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -22.42 |
| Energy contribution | -24.40 |
| Covariance contribution | 1.98 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.528108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9831688 120 - 20766785 UUGAUCGGAUCGUCGCUGUUGAUAAUGGUACAGUUAAUUUGGUAAAUUAUUUUGCAUUUUAGAAGCAACCCAAUCCAUUUUCCAACGUGUGGCAUUCACUGACACACAGUGGAGCAGUGG ...........((((((((((..(((((....(((..(((.(((((....)))))......)))..))).....)))))...))))).)))))...(((((.(((...)))...))))). ( -27.10) >DroSec_CAF1 69183 101 - 1 UUGAUCGGAUCGUCGCUGUUGAUAAUGGUACCGUUAAUUUGGUAAAUUAUUUUGCAUUUUAGAAGCAAACCA-----------AACGUGUGGCAUUCACUGACACACAGUG--------G ...........(((((((((((((((..(((((......))))).))))))((((.........))))....-----------)))).)))))...(((((.....)))))--------. ( -23.50) >DroEre_CAF1 74155 120 - 1 UUGAUCGGAUCGUCGCUGUUGAUAAUGGUACCGUUAAUUUGGUAAAUUAUUUUGCAUUUCAGACGCAACCCAAUCCAUUUUCCAACGUGUGGCAUUUACUGCCACACAGUGGAGCAGUGG ......((..((((..(((.((((((..(((((......))))).))))))..))).....))))....))...((((((((((..((((((((.....))))))))..))))).))))) ( -40.00) >DroYak_CAF1 71526 120 - 1 UUGAUCGGAUCGUCGCUGUUGAUAAUGGUACCGUUAAUUUGGUAAAUUAUUUUGCAUUUUAGACGCAACCCAAUCCAUUUUCCAACGUGUGGCAUUCACUGCCACACAGUGGAGCAGUGG ......((..((((..(((.((((((..(((((......))))).))))))..))).....))))....))...((((((((((..((((((((.....))))))))..))))).))))) ( -40.00) >DroAna_CAF1 68871 109 - 1 UUGAUCGGAUCGUCGCUGUUGAUAAUGGUACCGUUAAUUUGGUAAAUUAUUUUGCAUUUUAGACGCAAACCAAUCCAUUUUCCAAUGUGUGGCAUUCACGGCCAUGCCC----------- ......((((((((..(((.((((((..(((((......))))).))))))..))).....)))).......))))..........(((((((.......)))))))..----------- ( -25.71) >consensus UUGAUCGGAUCGUCGCUGUUGAUAAUGGUACCGUUAAUUUGGUAAAUUAUUUUGCAUUUUAGACGCAACCCAAUCCAUUUUCCAACGUGUGGCAUUCACUGCCACACAGUGGAGCAGUGG ......((..((((..(((.((((((..(((((......))))).))))))..))).....))))....)).........((((..((((((((.....))))))))..))))....... (-22.42 = -24.40 + 1.98)

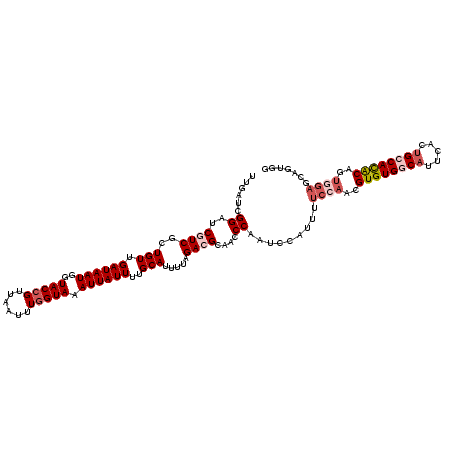

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:38 2006