
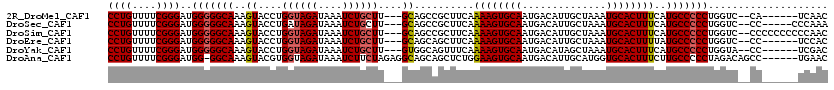
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,828,347 – 9,828,456 |
| Length | 109 |
| Max. P | 0.832678 |

| Location | 9,828,347 – 9,828,456 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -35.75 |
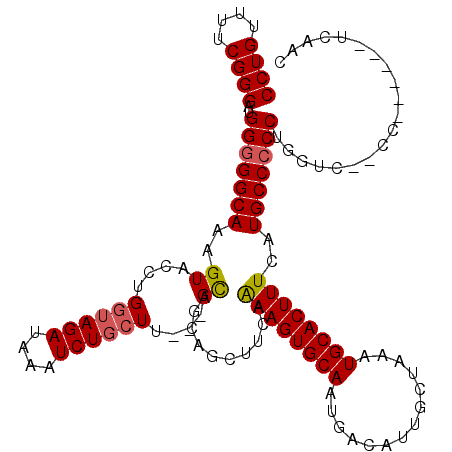
| Consensus MFE | -30.12 |
| Energy contribution | -30.34 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9828347 109 - 20766785 CCUGUUUUCGGGAUGGGGGCAAAGUACCUGGUAGAUAAAUCUGCUU---GCAGCCGCUUCAAAAGUGCAAUGACAUUGCUAAAUGCACUUUCAUGCCCCCUGGUC--CA------UCAAC .........(((..(((((((((((..((((((((....)))))..---.)))..))))..((((((((..............))))))))..)))))))...))--).------..... ( -34.54) >DroSec_CAF1 65989 110 - 1 CCUGUUUUCGGGAUGGGGGCAAAGUACCUGAUAGAUAAAUCUGCUU---GCAGCCGCUUCAAAAGUGCAAUGACAUUGCUAAAUGCACUUUCAUGCCCCCUGGUC--CC-----CCCAAA .........((((((((((((((((..(((.((((....))))...---.)))..))))..((((((((..............))))))))..)))))))..)))--))-----...... ( -34.94) >DroSim_CAF1 67783 115 - 1 CCUGUUUUCGGGAUGGGGGCAAAGUACCUGGUAGAUAAAUCUGCUU---GCAGCCGCUUCAAAAGUGCAAUGACAUUGCUAAAUGCACUUUCAUGCCCCCUGGUC--CCCCCCCCCCAAC .........((((((((((((((((..((((((((....)))))..---.)))..))))..((((((((..............))))))))..)))))))..)))--))........... ( -37.14) >DroEre_CAF1 70889 109 - 1 CCUGUUUUCGGGAUGGGGGCAAAGUACCUGGUAGAUAAAUCUGCUU---GCAGCAGCUUCAAAAGUGCAAUGACAUUGCUAAAUGCACUUUUAUGCCCCCUGGUC--CC------UCCAC .........((((((((((((((((..((((((((....)))))..---.)))..)))).(((((((((..............))))))))).)))))))..)))--))------..... ( -38.44) >DroYak_CAF1 68074 109 - 1 CCUGUUUUCGGGAUGGGGGCAAAGUACCUGGUAGAUAAAUCUGCUU---GUGGCAGUUUCAAAAGUGCAAUGACAUAGCUAAAUGCACUUUCAUGCCCCCUGGUA--CC------UCGAC ((((....))))..(((((((..((....((((((....)))))).---...)).......((((((((..............))))))))..))))))).....--..------..... ( -34.04) >DroAna_CAF1 66025 113 - 1 CCUGUUUUCGGGAUGG-GGCAAAGUACGUGGUAGAUAAAUCUUCUAGAGGCAGCAGCUCUGGAAGUGCAAUGACAUUGCAUGGUGCACUUUCUUGCCCCCUAGACAGCC------UGAAC ......((((((..((-(((((.(...(((..........(((((((((.......)))))))))((((((...)))))).....)))...))))))))........))------)))). ( -35.40) >consensus CCUGUUUUCGGGAUGGGGGCAAAGUACCUGGUAGAUAAAUCUGCUU___GCAGCAGCUUCAAAAGUGCAAUGACAUUGCUAAAUGCACUUUCAUGCCCCCUGGUC__CC______UCAAC ((((....))))..(((((((..((....((((((....))))))....))..........((((((((..............))))))))..))))))).................... (-30.12 = -30.34 + 0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:33 2006