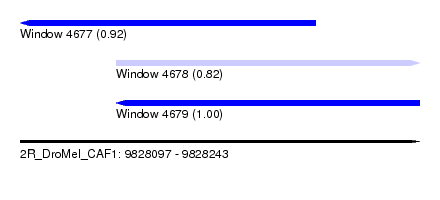
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,828,097 – 9,828,243 |
| Length | 146 |
| Max. P | 0.998080 |

| Location | 9,828,097 – 9,828,205 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.22 |
| Mean single sequence MFE | -23.65 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.68 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9828097 108 - 20766785 GCCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAAAUGCAAAAUGACAAAAAA-----AAACACAACAAAUAAUG (((....)))......(((((((..((((((((((........))))))))))...)))))))....((((..........))))......-----................. ( -22.70) >DroSec_CAF1 65731 113 - 1 GCCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAAAUACAAAAUGACAAAAAAAUAAGAAACACAACAAAUACUG (((....)))..(((((((((((..((((((((((........))))))))))...)))))))....((((..........))))..........)))).............. ( -24.40) >DroSim_CAF1 67550 112 - 1 -CCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAAAUGCAAAAUGACAAAAAAAUAAGAAACACAACAAAUAAUG -...(((((..((((.((....)).((((((((((........))))))))))....))))..))..((((..........))))..........)))............... ( -20.60) >DroYak_CAF1 67826 111 - 1 GCCAUUCGGCUU-UUCGAUUUUUCUGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAAAUGCAAAACG-CAAAAUGACAAAAAACACAACAAAUAAUG (((....)))..-...(((((((..((((((((((........))))))))))...)))))))....((((...(((.....)-))...)))).................... ( -26.90) >consensus GCCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAAAUGCAAAAUGACAAAAAAAUAAGAAACACAACAAAUAAUG (((....)))......(((((((..((((((((((........))))))))))...)))))))....((((..........))))............................ (-18.92 = -19.68 + 0.75)

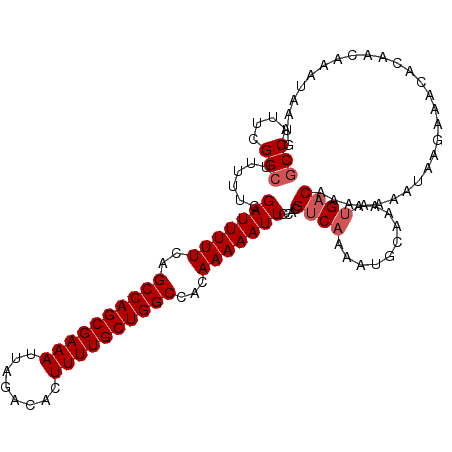
| Location | 9,828,132 – 9,828,243 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -29.17 |
| Consensus MFE | -22.25 |
| Energy contribution | -22.75 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9828132 111 + 20766785 UUUGACGUGGAAUUUUUGUGGCCAGCAAAAGUGUCUAAUUUCGCUGGCUGAAAAAUCGAAAAAGCCGAAUGGC-CGUAUAUAUAGCCACUUCAACGGC-AAUCACGCAGUUGU .(((.((((((.(((((..(((((((.(((........))).)))))))))))).))......((((..((((-..(....)..))))......))))-...))))))).... ( -31.50) >DroSec_CAF1 65771 113 + 1 UUUGACGUGGAAUUUUUGUGGCCAGCAAAAGUGUCUAAUUUCGCUGGCUGAAAAAUCGAAAAAGCCGAAUGGCCCGUAUGUAUAGCCACUUCAACAGCUAAACACGCAGUUGU ..(((.((((.((((((.((((((((.(((........))).)))))))))))))).......(((....)))............)))).)))((((((........)))))) ( -29.80) >DroSim_CAF1 67590 88 + 1 UUUGACGUGGAAUUUUUGUGGCCAGCAAAAGUGUCUAAUUUCGCUGGCUGAAAAAUCGAAAAAGCCGAAUGG-------------------------CUAAACACGCAGUUGU .(((.((((((.(((((..(((((((.(((........))).)))))))))))).)).....((((....))-------------------------))...))))))).... ( -26.50) >DroYak_CAF1 67865 106 + 1 UUUGACGUGGAAUUUUUGUGGCCAGCAAAAGUGUCUAAUUUCGCUGGCAGAAAAAUCGAA-AAGCCGAAUGGCC-----AAAAAGCCACUUGAACAGC-AAUCACGCAGUUUU .(((.((((((.(((((...((((((.(((........))).)))))).))))).))...-..(((((.((((.-----.....)))).)))....))-...))))))).... ( -28.90) >consensus UUUGACGUGGAAUUUUUGUGGCCAGCAAAAGUGUCUAAUUUCGCUGGCUGAAAAAUCGAAAAAGCCGAAUGGC______AUAUAGCCACUUCAACAGC_AAACACGCAGUUGU .(((.((((((.(((((..(((((((.(((........))).)))))))))))).))......(((....))).............................))))))).... (-22.25 = -22.75 + 0.50)

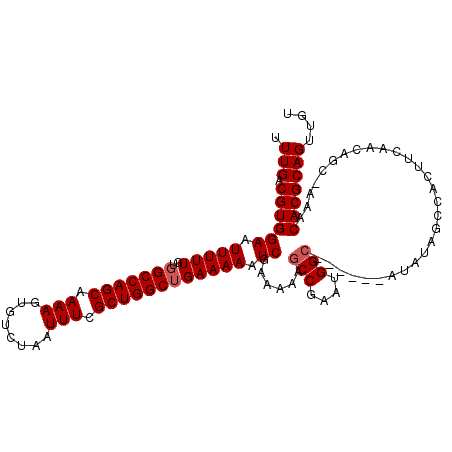
| Location | 9,828,132 – 9,828,243 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Mean single sequence MFE | -32.55 |
| Consensus MFE | -25.44 |
| Energy contribution | -27.38 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9828132 111 - 20766785 ACAACUGCGUGAUU-GCCGUUGAAGUGGCUAUAUAUACG-GCCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAA ......(((((...-((((....(((((((........)-))))))))))......(((((((..((((((((((........))))))))))...))))))).))))).... ( -36.00) >DroSec_CAF1 65771 113 - 1 ACAACUGCGUGUUUAGCUGUUGAAGUGGCUAUACAUACGGGCCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAA ....(((..(((.((((..(....)..)))).)))..)))(((....)))......(((((((..((((((((((........))))))))))...))))))).......... ( -33.70) >DroSim_CAF1 67590 88 - 1 ACAACUGCGUGUUUAG-------------------------CCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAA ......(((((...((-------------------------((....)))).....(((((((..((((((((((........))))))))))...))))))).))))).... ( -26.20) >DroYak_CAF1 67865 106 - 1 AAAACUGCGUGAUU-GCUGUUCAAGUGGCUUUUU-----GGCCAUUCGGCUU-UUCGAUUUUUCUGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAA ......(((((...-((((....(((((((....-----)))))))))))..-...(((((((..((((((((((........))))))))))...))))))).))))).... ( -34.30) >consensus ACAACUGCGUGAUU_GCUGUUGAAGUGGCUAUAU______GCCAUUCGGCUUUUUCGAUUUUUCAGCCAGCGAAAUUAGACACUUUUGCUGGCCACAAAAAUUCCACGUCAAA ......(((((....((((....((((((...........))))))))))......(((((((..((((((((((........))))))))))...))))))).))))).... (-25.44 = -27.38 + 1.94)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:32 2006