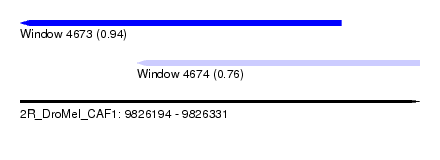
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,826,194 – 9,826,331 |
| Length | 137 |
| Max. P | 0.939770 |

| Location | 9,826,194 – 9,826,304 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.81 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -18.96 |
| Energy contribution | -19.10 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9826194 110 - 20766785 GUAUCUGUUGUAUGGAUAUGGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUUAGCGAAUUCCUCGCACUCUUUGCAAUGCCAGUACUAAUUU ((((((((..((....))..))))))))..........((((---------...((((((.(((-((........))))).((((.....))))..........))))))))))...... ( -32.40) >DroSec_CAF1 63900 104 - 1 GUAUCUGUUGUAUGAAUAUGGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUUAGCGAAUUCC------UCUUUGCAAUGCCAGUACUAAUUU ((((((((..((....))..))))))))..........((((---------...((((((.(((-((........))))).(((((....------..))))).))))))))))...... ( -31.20) >DroSim_CAF1 65426 104 - 1 GUAUCUGUUGUAUGAAUAUGGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUUAGCGAAUUCC------UCUUUGCAAUGCCAGUACUAAUUU ((((((((..((....))..))))))))..........((((---------...((((((.(((-((........))))).(((((....------..))))).))))))))))...... ( -31.20) >DroEre_CAF1 68728 110 - 1 GUAUCUGGUCUAUGGAUACAGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUUAGCGAAUUCCUCGCACUCUAUGCAAUGCCUGUACGAAUUU ((((((((((....)))....))))))).........(((((---------(.((.((((((((-((........)))...((((.....))))..)))))))...)).))))))..... ( -32.10) >DroYak_CAF1 65894 110 - 1 GUAUCUGCUGUAUGGAUACAGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUUAGCGAAUUCCUCGCACUCUAUGCAAUGGCUGUACACAUUU ((((((((((((....))))))))))))..........((((---------(.((.((((((((-((........)))...((((.....))))..)))))))....)))))))...... ( -38.80) >DroAna_CAF1 63898 119 - 1 GUAUCUUCCACCUGGGAGCGACAGAUACGGAUACAGAUACAGAUGGCGGCGGAGUAGUAUGGGGACACAAAUAUUUGUUUAGCGAAUUCCUAGCACUUUGUGGAAGUUAAAUACGAAA-U (((((((((((((((((((((((((((((....((........))....))........((....))....))))))))..))....))))))......)))))))....))))....-. ( -26.10) >consensus GUAUCUGUUGUAUGGAUACGGCAGAUACAAAUACACACGUAC_________AAGUGGUAUGGAG_CACAAAUAUUUGCUUAGCGAAUUCCUCGCACUCUUUGCAAUGCCAGUACUAAUUU ((((((((((((....))))))))))))..((((.(((...............)))))))((((........((((((...))))))........))))..................... (-18.96 = -19.10 + 0.14)

| Location | 9,826,234 – 9,826,331 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.64 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -17.39 |
| Energy contribution | -17.48 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9826234 97 - 20766785 -----CAA--UAAGAU---G---CUGGCCCAAGGUGGAAAGUAUCUGUUGUAUGGAUAUGGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUU -----...--..((((---(---.((((((..........((((((((..((....))..))))))))..((((.(((....---------..))))))))).)-).))..))))).... ( -25.40) >DroSec_CAF1 63934 100 - 1 -----CAA--UGAGAU---GCUGCUGGCCCAAGGUGGAAAGUAUCUGUUGUAUGAAUAUGGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUU -----...--....((---((..((........(((....((((((((..((....))..)))))))).....)))......---------.))..)))).(((-((........))))) ( -27.86) >DroSim_CAF1 65460 100 - 1 -----CAA--UGAGAU---GCUGCUGGCCCAAGGUGGAAAGUAUCUGUUGUAUGAAUAUGGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUU -----...--....((---((..((........(((....((((((((..((....))..)))))))).....)))......---------.))..)))).(((-((........))))) ( -27.86) >DroEre_CAF1 68768 97 - 1 -----CAG--UGAGAU---G---CUGGCCCAAGGUGGAAAGUAUCUGGUCUAUGGAUACAGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUU -----..(--((((((---(---((..((......))..))))))).((((.((....))..)))).......))).(((((---------.....)))))(((-((........))))) ( -24.60) >DroYak_CAF1 65934 97 - 1 -----CAG--UGAGAU---G---CUGGCCCAAGGUGGAAAGUAUCUGCUGUAUGGAUACAGCAGAUACAAAUACACACGUAC---------AAGUGGUAUGGAG-CACAAAUAUUUGCUU -----(((--(.....---)---)))((((..........((((((((((((....))))))))))))..((((.(((....---------..))))))))).)-).............. ( -32.70) >DroAna_CAF1 63937 120 - 1 CUUAUCAUUCCGCGAUGCUGCUGCUGCUCCAAGGUAGAAAGUAUCUUCCACCUGGGAGCGACAGAUACGGAUACAGAUACAGAUGGCGGCGGAGUAGUAUGGGGACACAAAUAUUUGUUU ..(((.(((((((..((((.(((.((((((.((((.(((......))).)))).)))))).)))....(....)..........))))))))))).)))((....))............. ( -36.00) >consensus _____CAA__UGAGAU___G___CUGGCCCAAGGUGGAAAGUAUCUGUUGUAUGGAUACGGCAGAUACAAAUACACACGUAC_________AAGUGGUAUGGAG_CACAAAUAUUUGCUU ...............................(((..((..((((((((((((....))))))))))))..((((.(((...............))))))).............))..))) (-17.39 = -17.48 + 0.09)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:27 2006