
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,825,595 – 9,825,701 |
| Length | 106 |
| Max. P | 0.543910 |

| Location | 9,825,595 – 9,825,701 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.47 |
| Mean single sequence MFE | -26.86 |
| Consensus MFE | -14.97 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9825595 106 + 20766785 GCG---AAUUUGGGC--UGAGAUCUGUUUCGCUGGAGCUGUAAAUUCCUC-ACAUAGUAAGCUGAAAUUUCGGGAAAACAACAUGCAGAGUGUUUUCCCAA------AUUUCGCAAAA-- (((---(((((.(((--(.....((((......((((.......))))..-..))))..)))).)))....(((((((((.(.......))))))))))..------..)))))....-- ( -27.60) >DroSec_CAF1 63308 103 + 1 GCG---AAUUU------UGGGAUCUGUUUCGCUGGAGCUGUAAGUUCCUCAACAUUGUAAGCUGAAAUUUCGGGAAAACAACAUGCAGAGUGUUUUCCCAA------AUUUCGCUAAA-- (((---((...------..........))))).(((((.....)))))...........(((.(((((((.(((((((((.(.......))))))))))))------))))))))...-- ( -30.72) >DroSim_CAF1 64862 103 + 1 GCG---AAUUU------UGGGAUCUGUUUCGCUGGAGCUGUAAAUUCCUCAACAUAGUAAGCUGAAAUUUCGGGAAAACAACAUGCAGAGUGUUUUCCCAA------AUUUCGCUAAA-- (((---((.((------(..((...((((((((...(((((............))))).))).))))).))(((((((((.(.......))))))))))))------).)))))....-- ( -27.50) >DroEre_CAF1 68103 95 + 1 ------------------CAGAUCUGUGUCGUUGGAGUUGUAAAUUCCUC-ACAUAAUAAGCUGAAAUUUCGGGAAAGCAACAUGCAGAGUGUUUUCCCAA------AUUUCGCAAAAAA ------------------......(((((.(..(((((.....))))).)-)))))....((.(((((((.(((((((((.(.......))))))))))))------)))))))...... ( -25.90) >DroYak_CAF1 65220 108 + 1 GCG---AAUUUGGAC--UCAGAUCUGUUUCGGUGGAGCUGUAAAUUCCUC-ACAUAAUAAGCUGAAAUUUCGGGAAAGCAACAUGCACAGUGUUUUCCCAA------AUUUCGCAAAAAA (((---((((((.((--((..(((......))).))).).)))))))...-............(((((((.(((((((((.(.......))))))))))))------)))))))...... ( -28.50) >DroAna_CAF1 63338 118 + 1 GAUUCUGUUCUGGGAUAUGAGAUCUGAGAUCUGGGAGCUGUAAAUUUCUC-ACACAUUAAGCUGAUUUUUCGGGAAA-UAACCUGCAAAGUAUUUUCCCAAAAAAAAAUUGCGCAAAAUA .(((.((((((.((((.....)))).)))..((((((((...........-........)))..(((((.((((...-...)))).)))))....)))))............))).))). ( -20.91) >consensus GCG___AAUUU______UGAGAUCUGUUUCGCUGGAGCUGUAAAUUCCUC_ACAUAGUAAGCUGAAAUUUCGGGAAAACAACAUGCAGAGUGUUUUCCCAA______AUUUCGCAAAA__ ....................((...(((((.....((((....................))))))))).))(((((((((.(.......))))))))))..................... (-14.97 = -14.25 + -0.72)

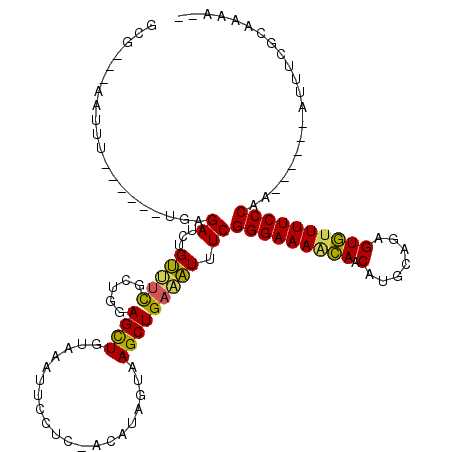
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:25 2006