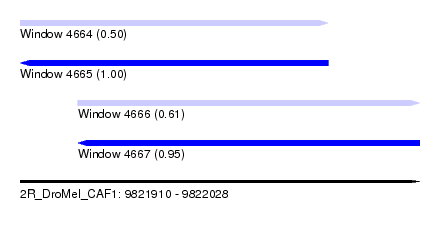
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,821,910 – 9,822,028 |
| Length | 118 |
| Max. P | 0.999431 |

| Location | 9,821,910 – 9,822,001 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -21.68 |
| Consensus MFE | -18.96 |
| Energy contribution | -18.96 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.87 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9821910 91 + 20766785 GAGUCCUCAAGAACUCAGUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUGCUUUCCUCAUUCGUUGGCUCUUUU------ (((((..(.(((((...)))))(((((...(((...((((((...........)))))).)))....))))).......)..)))))....------ ( -20.40) >DroSec_CAF1 59652 90 + 1 GAGUCCUCAAGAACUCAGUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUACUUUCCUCAUUCGCUGGCUCUU-U------ (((((..(.(((((...)))))(((((...(((...((((((...........)))))).)))....))))).......)..)))))..-.------ ( -18.30) >DroSim_CAF1 61148 90 + 1 GAGUCCUCGAGAACUCAGUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUACUUUCCUCAUUCGCUGGCUCUU-U------ (((((..((((...(((((((....)))))))....((((((...........)))))).................))))..)))))..-.------ ( -21.30) >DroEre_CAF1 64360 97 + 1 GAGUCCUCAAGAACUCAGUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUGCUUUCCUCAUUCGCUGGCUCUUUUGGCUGU .((.((..((((.(.((((...(((((...(((...((((((...........)))))).)))....))))).......))))).))))..)))).. ( -22.60) >DroYak_CAF1 61456 97 + 1 GAGUCCUCAAGAACUCAGUUUUGAAAGGCUGGACGCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUGCUUUCCUCAUUCGCUGGCUCUUUUGGCUGU .((.((..((((.(.((((..((((((((.(((...((((((...........)))))).)))...)))))..)))...))))).))))..)))).. ( -25.80) >consensus GAGUCCUCAAGAACUCAGUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUGCUUUCCUCAUUCGCUGGCUCUUUU______ (((((..(.(((((...)))))(((((...(((...((((((...........)))))).)))....))))).......)..))))).......... (-18.96 = -18.96 + 0.00)

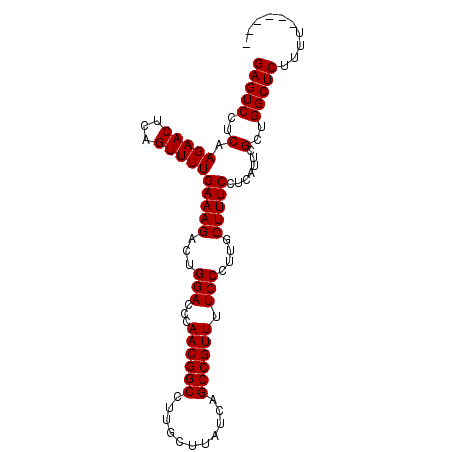
| Location | 9,821,910 – 9,822,001 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.27 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -25.14 |
| Energy contribution | -25.54 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.92 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9821910 91 - 20766785 ------AAAAGAGCCAACGAAUGAGGAAAGCAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAACUGAGUUCUUGAGGACUC ------...................(((((.(..(((.(((((((.........)))))))...)))..))))))......((((((....)))))) ( -24.90) >DroSec_CAF1 59652 90 - 1 ------A-AAGAGCCAGCGAAUGAGGAAAGUAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAACUGAGUUCUUGAGGACUC ------.-(((((((((........(((((....(((.(((((((.........)))))))...)))...)))))....))).))))))........ ( -27.00) >DroSim_CAF1 61148 90 - 1 ------A-AAGAGCCAGCGAAUGAGGAAAGUAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAACUGAGUUCUCGAGGACUC ------.-..(((((.(.((((...(((((....(((.(((((((.........)))))))...)))...)))))........)))).)..)).))) ( -25.60) >DroEre_CAF1 64360 97 - 1 ACAGCCAAAAGAGCCAGCGAAUGAGGAAAGCAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAACUGAGUUCUUGAGGACUC ..((((..(((((((((........(((((.(..(((.(((((((.........)))))))...)))..))))))....))).))))))..)).)). ( -30.10) >DroYak_CAF1 61456 97 - 1 ACAGCCAAAAGAGCCAGCGAAUGAGGAAAGCAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGCGUCCAGCCUUUCAAAACUGAGUUCUUGAGGACUC ..((((..(((((((((........(((((....(((.(((((((.........)))))))...)))...)))))....))).))))))..)).)). ( -29.70) >consensus ______AAAAGAGCCAGCGAAUGAGGAAAGCAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAACUGAGUUCUUGAGGACUC ........(((((((((........(((((....(((.(((((((.........)))))))...)))...)))))....))).))))))........ (-25.14 = -25.54 + 0.40)


| Location | 9,821,927 – 9,822,028 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -19.30 |
| Consensus MFE | -17.41 |
| Energy contribution | -17.30 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9821927 101 + 20766785 GUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUGCUUUCCUCAUUCGUUGGCUCUUUUAGACGCACGAUCUUCUUAUUCCC-UUUU .....(((((...(((...((((((...........)))))).)))....))))).....((((..((((.....)).)))))).............-.... ( -17.90) >DroSec_CAF1 59669 101 + 1 GUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUACUUUCCUCAUUCGCUGGCUCUU-UGGCCACACGAUCCUCUUAUUCCCCUUUU .....(((((...(((...((((((...........)))))).)))....))))).....(((.(((((...-.)))))..))).................. ( -20.00) >DroSim_CAF1 61165 101 + 1 GUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUACUUUCCUCAUUCGCUGGCUCUU-UGGCCACACGAUCUUCUUAUUCCCCUUUU .....(((((...(((...((((((...........)))))).)))....))))).....(((.(((((...-.)))))..))).................. ( -20.00) >consensus GUUUUGAAAGACUGGACCCAACGGCCUUGCUUAUCAGCCGUUUUCCCUUACUUUCCUCAUUCGCUGGCUCUU_UGGCCACACGAUCUUCUUAUUCCCCUUUU .....(((((...(((...((((((...........)))))).)))....))))).....(((.(((((.....)))))..))).................. (-17.41 = -17.30 + -0.11)

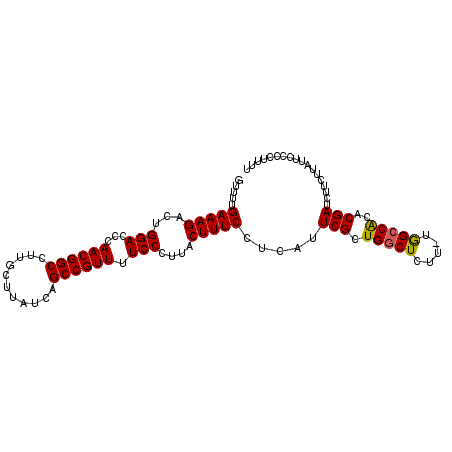
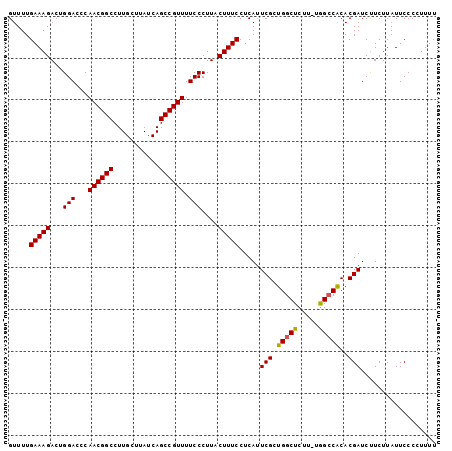
| Location | 9,821,927 – 9,822,028 |
|---|---|
| Length | 101 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 94.75 |
| Mean single sequence MFE | -27.07 |
| Consensus MFE | -24.52 |
| Energy contribution | -24.97 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 9821927 101 - 20766785 AAAA-GGGAAUAAGAAGAUCGUGCGUCUAAAAGAGCCAACGAAUGAGGAAAGCAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAAC ....-.............((((((.((.....))))..)))).....(((((.(..(((.(((((((.........)))))))...)))..))))))..... ( -23.80) >DroSec_CAF1 59669 101 - 1 AAAAGGGGAAUAAGAGGAUCGUGUGGCCA-AAGAGCCAGCGAAUGAGGAAAGUAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAAC ..................((((.((((..-....)))))))).....(((((....(((.(((((((.........)))))))...)))...)))))..... ( -28.70) >DroSim_CAF1 61165 101 - 1 AAAAGGGGAAUAAGAAGAUCGUGUGGCCA-AAGAGCCAGCGAAUGAGGAAAGUAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAAC ..................((((.((((..-....)))))))).....(((((....(((.(((((((.........)))))))...)))...)))))..... ( -28.70) >consensus AAAAGGGGAAUAAGAAGAUCGUGUGGCCA_AAGAGCCAGCGAAUGAGGAAAGUAAGGGAAAACGGCUGAUAAGCAAGGCCGUUGGGUCCAGUCUUUCAAAAC ..................((((.((((.......)))))))).....(((((....(((.(((((((.........)))))))...)))...)))))..... (-24.52 = -24.97 + 0.45)

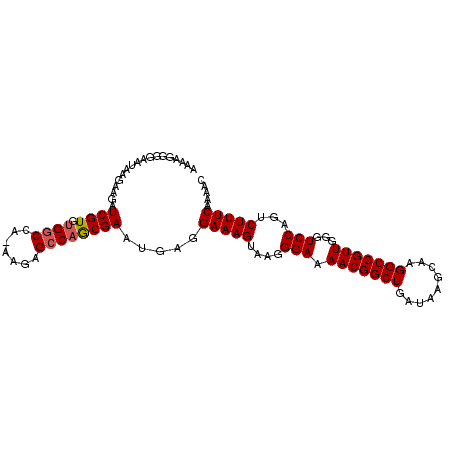

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:20 2006