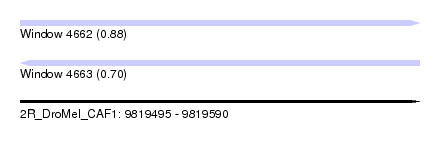
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 9,819,495 – 9,819,590 |
| Length | 95 |
| Max. P | 0.883966 |

| Location | 9,819,495 – 9,819,590 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -28.12 |
| Consensus MFE | -24.44 |
| Energy contribution | -24.38 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
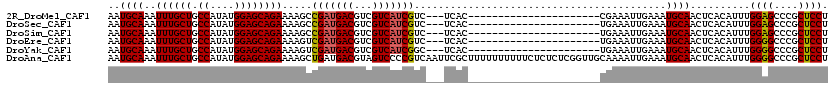
>2R_DroMel_CAF1 9819495 95 + 20766785 AAUGCAAAUUUGCUGCCAUAUGGAGCAGAAAAGCCGAUGACGUCGUCAUCGUC---UCAC----------------------CGAAAUUGAAAUGCAACUCACAUUUGGAGCCCGCUCCU ..((((..((((((.((....))))))))..((.(((((((...))))))).)---)...----------------------...........))))..........((((....)))). ( -26.70) >DroSec_CAF1 57340 95 + 1 AAUGCAAAUUUGCUGCCAUAUGGAGCAGAAAAGCCGAUGACGUCGUCAUCGUC---UCAC----------------------UGAAAUUGAAAUGCAACUCACAUUUGGAGCCCGCUCCU ..((((..((((((.((....))))))))..((.(((((((...))))))).)---)...----------------------...........))))..........((((....)))). ( -26.70) >DroSim_CAF1 58793 95 + 1 AAUGCAAAUUUGCUGCCAUAUGGAGCAGAAAAGCCGAUGACGUCGUCAUCGUC---UCAC----------------------UGAAAUUGAAAUGCAACUCACAUUUGGAGCCCGCUCCU ..((((..((((((.((....))))))))..((.(((((((...))))))).)---)...----------------------...........))))..........((((....)))). ( -26.70) >DroEre_CAF1 62090 95 + 1 AAUGCAAAUUUGCUGCCAUAUGGAGCAGAAAAGUCGAUGACGUCGUCAUCGUC---UCAC----------------------UGAAAUUGAAAUGCAACUCACAUUUGGGGCCCGCUCCU ..((((..((((((.((....))))))))..((.(((((((...))))))).)---)...----------------------...........))))..........((((....)))). ( -26.90) >DroYak_CAF1 59117 95 + 1 AAUGCAAAUUUGCUGCCAUAUGGAGCAGAAAAGUCGAUGACGUCGUCAUCGGC---UCAC----------------------UGAAAUUGAAAUGCAACUCACAUUUGGGGCCCGCUCCU ..((((..((((((.((....))))))))..((((((((((...)))))))))---)...----------------------...........))))..........((((....)))). ( -30.20) >DroAna_CAF1 57709 120 + 1 AAUGCAAAUUUGCUGCCAUAUGGAGCAGAAAAGCUGAUGACGUAGUCCCCGUCAAUUCGCUUUUUUUUUUCUCUCUCGGUUGCAAAAUUGAAAUGCAACUCACAUUUGGGGCCCGCUCCU ...(((....)))........(((((((((((((.(((((((.......)))).))).))))))))...........((((.((((..(((........)))..)))).)))).))))). ( -31.50) >consensus AAUGCAAAUUUGCUGCCAUAUGGAGCAGAAAAGCCGAUGACGUCGUCAUCGUC___UCAC______________________UGAAAUUGAAAUGCAACUCACAUUUGGAGCCCGCUCCU ..((((..((((((.((....)))))))).....(((((((...)))))))..........................................))))..........((((....)))). (-24.44 = -24.38 + -0.06)

| Location | 9,819,495 – 9,819,590 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -25.54 |
| Energy contribution | -25.73 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697600 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
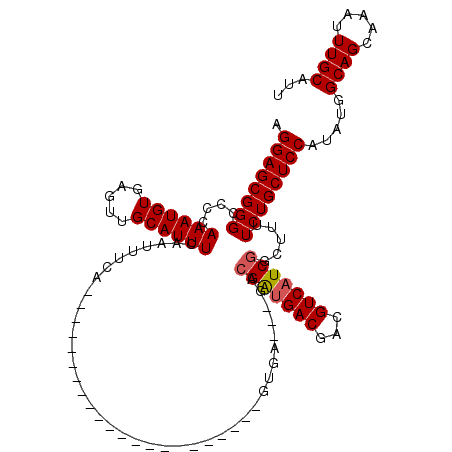
>2R_DroMel_CAF1 9819495 95 - 20766785 AGGAGCGGGCUCCAAAUGUGAGUUGCAUUUCAAUUUCG----------------------GUGA---GACGAUGACGACGUCAUCGGCUUUUCUGCUCCAUAUGGCAGCAAAUUUGCAUU .((((....)))).((((..((((((.(((((......----------------------.)))---))(((((((...))))))))).....((((((....)).))))))))..)))) ( -30.70) >DroSec_CAF1 57340 95 - 1 AGGAGCGGGCUCCAAAUGUGAGUUGCAUUUCAAUUUCA----------------------GUGA---GACGAUGACGACGUCAUCGGCUUUUCUGCUCCAUAUGGCAGCAAAUUUGCAUU .((((....)))).((((..((((((.(((((......----------------------.)))---))(((((((...))))))))).....((((((....)).))))))))..)))) ( -30.70) >DroSim_CAF1 58793 95 - 1 AGGAGCGGGCUCCAAAUGUGAGUUGCAUUUCAAUUUCA----------------------GUGA---GACGAUGACGACGUCAUCGGCUUUUCUGCUCCAUAUGGCAGCAAAUUUGCAUU .((((....)))).((((..((((((.(((((......----------------------.)))---))(((((((...))))))))).....((((((....)).))))))))..)))) ( -30.70) >DroEre_CAF1 62090 95 - 1 AGGAGCGGGCCCCAAAUGUGAGUUGCAUUUCAAUUUCA----------------------GUGA---GACGAUGACGACGUCAUCGACUUUUCUGCUCCAUAUGGCAGCAAAUUUGCAUU .((((((((....((((((.....))))))........----------------------..((---(.(((((((...))))))).))).)))))))).....((((.....))))... ( -27.90) >DroYak_CAF1 59117 95 - 1 AGGAGCGGGCCCCAAAUGUGAGUUGCAUUUCAAUUUCA----------------------GUGA---GCCGAUGACGACGUCAUCGACUUUUCUGCUCCAUAUGGCAGCAAAUUUGCAUU .((((((((....((((((.....))))))........----------------------..((---(.(((((((...))))))).))).)))))))).....((((.....))))... ( -28.30) >DroAna_CAF1 57709 120 - 1 AGGAGCGGGCCCCAAAUGUGAGUUGCAUUUCAAUUUUGCAACCGAGAGAGAAAAAAAAAAGCGAAUUGACGGGGACUACGUCAUCAGCUUUUCUGCUCCAUAUGGCAGCAAAUUUGCAUU .(((((((..........((.((((((.........))))))))............((((((....(((((.......)))))...))))))))))))).....((((.....))))... ( -34.70) >consensus AGGAGCGGGCCCCAAAUGUGAGUUGCAUUUCAAUUUCA______________________GUGA___GACGAUGACGACGUCAUCGGCUUUUCUGCUCCAUAUGGCAGCAAAUUUGCAUU .((((((((....((((((.....)))))).......................................(((((((...))))))).....)))))))).....((((.....))))... (-25.54 = -25.73 + 0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:40:16 2006